Post Syndicated from Abdel Jaidi original https://aws.amazon.com/blogs/big-data/advanced-patterns-with-aws-sdk-for-pandas-on-aws-glue-for-ray/
AWS SDK for pandas is a popular Python library among data scientists, data engineers, and developers. It simplifies interaction between AWS data and analytics services and pandas DataFrames. It allows easy integration and data movement between 22 types of data stores, including Amazon Simple Storage Service (Amazon S3), Amazon Athena, Amazon Redshift, and Amazon OpenSearch Service.
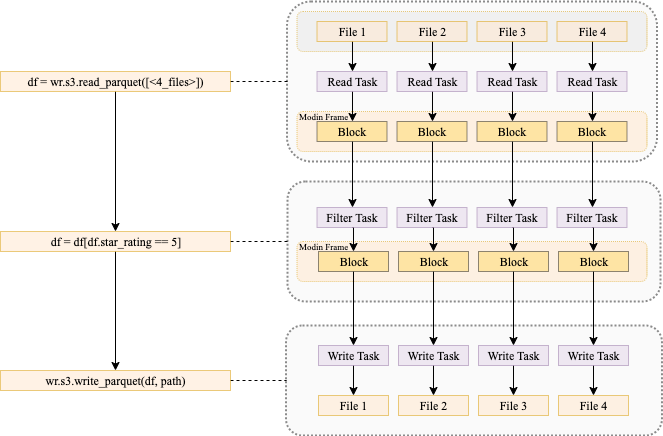
In the previous post, we discussed how you can use AWS SDK for pandas to scale your workloads on AWS Glue for Ray. We explained how using both Ray and Modin within the library enabled us to distribute workloads across a compute cluster. To illustrate these capabilities, we explored examples of writing Parquet files to Amazon S3 at scale and querying data in parallel with Athena.
In this post, we show some more advanced ways to use this library on AWS Glue for Ray. We cover features and APIs from AWS services such as S3 Select, Amazon DynamoDB, and Amazon Timestream.
Solution overview
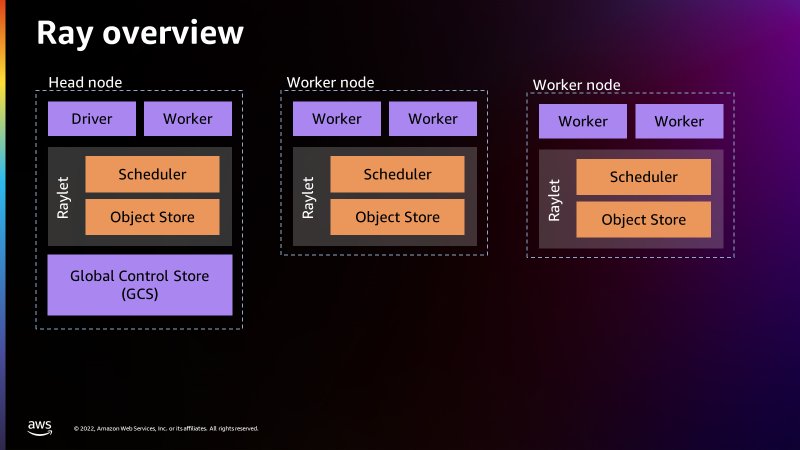
The Ray and Modin frameworks allow scaling of pandas workloads easily. You can write code on your laptop that uses the SDK for pandas to get data from an AWS data or analytics service to a pandas DataFrame, transform it using pandas, and then write it back to the AWS service. By using the distributed version of the SDK for pandas and replacing pandas with Modin, exactly the same code will scale on a Ray runtime—all logic about task coordination and distribution is hidden. Taking advantage of these abstractions, the AWS SDK for pandas team has made considerable use of Ray primitives to distribute some of the existing APIs (for the full list, see Supported APIs).
In this post, we show how to use some of these APIs in an AWS Glue for Ray job, namely querying with S3 Select, writing to and reading from a DynamoDB table, and writing to a Timestream table. Because AWS Glue for Ray is a fully managed environment, it’s by far the easiest way to run jobs because you don’t need to worry about cluster management. If you want to create your own cluster on Amazon Elastic Compute Cloud (Amazon EC2), refer to Distributing Calls on Ray Remote Cluster.
Configure solution resources
We use an AWS CloudFormation stack to provision the solution resources. Complete the following steps:
- Choose Launch stack to provision the stack in your AWS account:
This takes about 2 minutes to complete. On successful deployment, the CloudFormation stack shows the status as CREATE_COMPLETE.

- Navigate to AWS Glue Studio to find an AWS Glue job named
AdvancedGlueRayJob.

- On the Job details tab, scroll down and choose Advanced Properties.
Under Job Parameters, AWS SDK for pandas is specified as an additional Python module to install, along with Modin as an extra dependency.

- To run the job, choose Run and navigate to the Runs tab to monitor the job’s progress.

Import the library
To import the library, use the following code:
import awswrangler as wr
AWS SDK for pandas detects if the runtime supports Ray, and automatically initializes a cluster with the default parameters. Advanced users can override this process by starting the Ray runtime before the import command.
Scale S3 Select workflows
S3 Select allows you to use SQL statements to query and filter S3 objects, including compressed files. This can be particularly useful if you have large files of several TBs and want to extract some information. Because the workload is delegated to Amazon S3, you don’t have to download and filter objects on the client side, leading to lower latency, lower cost, and higher performance.
With AWS SDK for pandas, these calls to S3 Select can be distributed across Ray workers in the cluster. In the following example, we query Amazon reviews data in Parquet format, filtering for reviews with 5-star ratings in the Mobile_Electronics partition. star_rating is a column in the Parquet data itself, while the partition is a directory.
scan_range_chunk_size is an important parameter to calibrate when using S3 Select. It specifies the range of bytes to query the S3 object, thereby determining the amount of work delegated to each worker. For this example, it’s set to 16 MB, meaning the work of scanning the object is parallelized into separate S3 Select requests each 16 MB in size. A higher value equates to larger chunks per worker but fewer workers, and vice versa.
The results are returned in a Modin DataFrame, which is a drop-in replacement for pandas. It exposes the same APIs but enables you to use all the workers in the cluster. The data in the Modin DataFrame is distributed along with all the operations among the workers.
Scale DynamoDB workflows
DynamoDB is a scalable NoSQL database service that provides high-performance, low-latency, and managed storage.
AWS SDK for pandas uses Ray to scale DynamoDB workflows, allowing parallel data retrieval and insertion operations. The wr.dynamodb.read_items function retrieves data from DynamoDB in parallel across multiple workers, and the results are returned as a Modin DataFrame. Similarly, data insertion into DynamoDB can be parallelized using the wr.dynamodb.put_df function.
For example, the following code inserts the Amazon Reviews DataFrame obtained from S3 Select into a DynamoDB table and then reads it back:
DynamoDB calls are subject to AWS service quotas. The concurrency can be limited using the use_threads parameter.
Scale Timestream workflows
Timestream is a fast, scalable, fully managed, purpose-built time series database that makes it easy to store and analyze trillions of time series data points per day. With AWS SDK for pandas, you can distribute Timestream write operations across multiple workers in your cluster.
Data can be written to Timestream using the wr.timestream.write function, which parallelizes the data insertion process for improved performance.
In this example, we use sample data from Amazon S3 loaded into a Modin DataFrame. Familiar pandas commands such as selecting columns or resetting the index are applied at scale with Modin:
The Timestream write operation is parallelized across blocks in your dataset. If the blocks are too big, you can use Ray to repartition the dataset and increase the throughput, because each block will be handled by a separate thread:
We are now ready to insert the data into Timestream, and a final query confirms the number of rows in the table:
Clean up
To prevent unwanted charges to your AWS account, we recommend deleting the AWS resources that you used in this post:
- On the Amazon S3 console, empty the data from the S3 bucket with prefix
glue-ray-blog-script.

- On the AWS CloudFormation console, delete the
AdvancedSDKPandasOnGlueRaystack.
All resources will be automatically deleted with it.
Conclusion
In this post, we showcased some more advanced patterns to run your workloads using AWS SDK for pandas. In particular, these examples demonstrated how Ray is used within the library to distribute operations for several other AWS services, not just Amazon S3. When used in combination with AWS Glue for Ray, this gives you access to a fully managed environment to run at scale. We hope this solution can help with migrating your existing pandas jobs to achieve higher performance and speedups across multiple data stores on AWS.
About the Authors
 Abdel Jaidi is a Senior Cloud Engineer for AWS Professional Services. He works on open-source projects focused on AWS Data & Analytics services. In his spare time, he enjoys playing tennis and hiking.
Abdel Jaidi is a Senior Cloud Engineer for AWS Professional Services. He works on open-source projects focused on AWS Data & Analytics services. In his spare time, he enjoys playing tennis and hiking.
 Anton Kukushkin is a Data Engineer for AWS Professional Services based in London, UK. In his spare time, he enjoys playing musical instruments.
Anton Kukushkin is a Data Engineer for AWS Professional Services based in London, UK. In his spare time, he enjoys playing musical instruments.
 Leon Luttenberger is a Data Engineer for AWS Professional Services based in Austin, Texas. He works on AWS open-source solutions that help our customers analyze their data at scale. In his spare time, he enjoys reading and traveling.
Leon Luttenberger is a Data Engineer for AWS Professional Services based in Austin, Texas. He works on AWS open-source solutions that help our customers analyze their data at scale. In his spare time, he enjoys reading and traveling.