Post Syndicated from Radhika Jakkula original https://aws.amazon.com/blogs/big-data/orchestrate-an-end-to-end-etl-pipeline-using-amazon-s3-aws-glue-and-amazon-redshift-serverless-with-amazon-mwaa/
Amazon Managed Workflows for Apache Airflow (Amazon MWAA) is a managed orchestration service for Apache Airflow that you can use to set up and operate data pipelines in the cloud at scale. Apache Airflow is an open source tool used to programmatically author, schedule, and monitor sequences of processes and tasks, referred to as workflows. With Amazon MWAA, you can use Apache Airflow and Python to create workflows without having to manage the underlying infrastructure for scalability, availability, and security.
By using multiple AWS accounts, organizations can effectively scale their workloads and manage their complexity as they grow. This approach provides a robust mechanism to mitigate the potential impact of disruptions or failures, making sure that critical workloads remain operational. Additionally, it enables cost optimization by aligning resources with specific use cases, making sure that expenses are well controlled. By isolating workloads with specific security requirements or compliance needs, organizations can maintain the highest levels of data privacy and security. Furthermore, the ability to organize multiple AWS accounts in a structured manner allows you to align your business processes and resources according to your unique operational, regulatory, and budgetary requirements. This approach promotes efficiency, flexibility, and scalability, enabling large enterprises to meet their evolving needs and achieve their goals.
This post demonstrates how to orchestrate an end-to-end extract, transform, and load (ETL) pipeline using Amazon Simple Storage Service (Amazon S3), AWS Glue, and Amazon Redshift Serverless with Amazon MWAA.
Solution overview
For this post, we consider a use case where a data engineering team wants to build an ETL process and give the best experience to their end-users when they want to query the latest data after new raw files are added to Amazon S3 in the central account (Account A in the following architecture diagram). The data engineering team wants to separate the raw data into its own AWS account (Account B in the diagram) for increased security and control. They also want to perform the data processing and transformation work in their own account (Account B) to compartmentalize duties and prevent any unintended changes to the source raw data present in the central account (Account A). This approach allows the team to process the raw data extracted from Account A to Account B, which is dedicated for data handling tasks. This makes sure the raw and processed data can be maintained securely separated across multiple accounts, if required, for enhanced data governance and security.
Our solution uses an end-to-end ETL pipeline orchestrated by Amazon MWAA that looks for new incremental files in an Amazon S3 location in Account A, where the raw data is present. This is done by invoking AWS Glue ETL jobs and writing to data objects in a Redshift Serverless cluster in Account B. The pipeline then starts running stored procedures and SQL commands on Redshift Serverless. As the queries finish running, an UNLOAD operation is invoked from the Redshift data warehouse to the S3 bucket in Account A.
Because security is important, this post also covers how to configure an Airflow connection using AWS Secrets Manager to avoid storing database credentials within Airflow connections and variables.
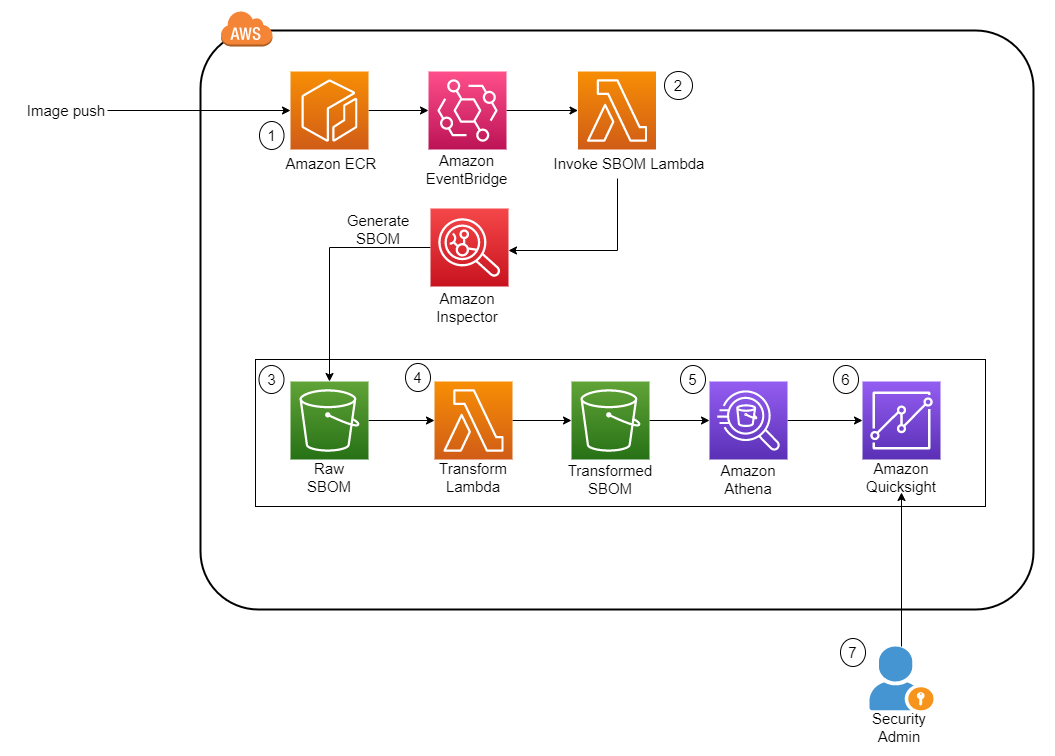
The following diagram illustrates the architectural overview of the components involved in the orchestration of the workflow.

The workflow consists of the following components:
- The source and target S3 buckets are in a central account (Account A), whereas Amazon MWAA, AWS Glue, and Amazon Redshift are in a different account (Account B). Cross-account access has been set up between S3 buckets in Account A with resources in Account B to be able to load and unload data.
- In the second account, Amazon MWAA is hosted in one VPC and Redshift Serverless in a different VPC, which are connected through VPC peering. A Redshift Serverless workgroup is secured inside private subnets across three Availability Zones.
- Secrets like user name, password, DB port, and AWS Region for Redshift Serverless are stored in Secrets Manager.
- VPC endpoints are created for Amazon S3 and Secrets Manager to interact with other resources.
- Usually, data engineers create an Airflow Directed Acyclic Graph (DAG) and commit their changes to GitHub. With GitHub actions, they are deployed to an S3 bucket in Account B (for this post, we upload the files into S3 bucket directly). The S3 bucket stores Airflow-related files like DAG files,
requirements.txtfiles, and plugins. AWS Glue ETL scripts and assets are stored in another S3 bucket. This separation helps maintain organization and avoid confusion. - The Airflow DAG uses various operators, sensors, connections, tasks, and rules to run the data pipeline as needed.
- The Airflow logs are logged in Amazon CloudWatch, and alerts can be configured for monitoring tasks. For more information, see Monitoring dashboards and alarms on Amazon MWAA.
Prerequisites
Because this solution centers around using Amazon MWAA to orchestrate the ETL pipeline, you need to set up certain foundational resources across accounts beforehand. Specifically, you need to create the S3 buckets and folders, AWS Glue resources, and Redshift Serverless resources in their respective accounts prior to implementing the full workflow integration using Amazon MWAA.
Deploy resources in Account A using AWS CloudFormation
In Account A, launch the provided AWS CloudFormation stack to create the following resources:
- The source and target S3 buckets and folders. As a best practice, the input and output bucket structures are formatted with hive style partitioning as
s3://<bucket>/products/YYYY/MM/DD/. - A sample dataset called
products.csv, which we use in this post.
Upload the AWS Glue job to Amazon S3 in Account B
In Account B, create an Amazon S3 location called aws-glue-assets-<account-id>-<region>/scripts (if not present). Replace the parameters for the account ID and Region in the sample_glue_job.py script and upload the AWS Glue job file to the Amazon S3 location.
Deploy resources in Account B using AWS CloudFormation
In Account B, launch the provided CloudFormation stack template to create the following resources:
- The S3 bucket
airflow-<username>-bucketto store Airflow-related files with the following structure:- dags – The folder for DAG files.
- plugins – The file for any custom or community Airflow plugins.
- requirements – The
requirements.txtfile for any Python packages. - scripts – Any SQL scripts used in the DAG.
- data – Any datasets used in the DAG.
- A Redshift Serverless environment. The name of the workgroup and namespace are prefixed with
sample. - An AWS Glue environment, which contains the following:
- An AWS Glue crawler, which crawls the data from the S3 source bucket
sample-inp-bucket-etl-<username>in Account A. - A database called
products_dbin the AWS Glue Data Catalog. - An ELT job called
sample_glue_job. This job can read files from theproductstable in the Data Catalog and load data into the Redshift tableproducts.
- An AWS Glue crawler, which crawls the data from the S3 source bucket
- A VPC gateway endpointto Amazon S3.
- An Amazon MWAA environment. For detailed steps to create an Amazon MWAA environment using the Amazon MWAA console, refer to Introducing Amazon Managed Workflows for Apache Airflow (MWAA).
Create Amazon Redshift resources
Create two tables and a stored procedure on an Redshift Serverless workgroup using the products.sql file.
In this example, we create two tables called products and products_f. The name of the stored procedure is sp_products.
Configure Airflow permissions
After the Amazon MWAA environment is created successfully, the status will show as Available. Choose Open Airflow UI to view the Airflow UI. DAGs are automatically synced from the S3 bucket and visible in the UI. However, at this stage, there are no DAGs in the S3 folder.
Add the customer managed policy AmazonMWAAFullConsoleAccess, which grants Airflow users permissions to access AWS Identity and Access Management (IAM) resources, and attach this policy to the Amazon MWAA role. For more information, see Accessing an Amazon MWAA environment.
The policies attached to the Amazon MWAA role have full access and must only be used for testing purposes in a secure test environment. For production deployments, follow the least privilege principle.
Set up the environment
This section outlines the steps to configure the environment. The process involves the following high-level steps:
- Update any necessary providers.
- Set up cross-account access.
- Establish a VPC peering connection between the Amazon MWAA VPC and Amazon Redshift VPC.
- Configure Secrets Manager to integrate with Amazon MWAA.
- Define Airflow connections.
Update the providers
Follow the steps in this section if your version of Amazon MWAA is less than 2.8.1 (the latest version as of writing this post).
Providers are packages that are maintained by the community and include all the core operators, hooks, and sensors for a given service. The Amazon provider is used to interact with AWS services like Amazon S3, Amazon Redshift Serverless, AWS Glue, and more. There are over 200 modules within the Amazon provider.
Although the version of Airflow supported in Amazon MWAA is 2.6.3, which comes bundled with the Amazon provided package version 8.2.0, support for Amazon Redshift Serverless was not added until the Amazon provided package version 8.4.0. Because the default bundled provider version is older than when Redshift Serverless support was introduced, the provider version must be upgraded in order to use that functionality.
The first step is to update the constraints file and requirements.txt file with the correct versions. Refer to Specifying newer provider packages for steps to update the Amazon provider package.
- Specify the requirements as follows:
- Update the version in the constraints file to 8.4.0 or higher.
- Add the constraints-3.11-updated.txt file to the
/dagsfolder.
Refer to Apache Airflow versions on Amazon Managed Workflows for Apache Airflow for correct versions of the constraints file depending on the Airflow version.
- Navigate to the Amazon MWAA environment and choose Edit.
- Under DAG code in Amazon S3, for Requirements file, choose the latest version.
- Choose Save.
This will update the environment and new providers will be in effect.
- To verify the providers version, go to Providers under the Admin table.
The version for the Amazon provider package should be 8.4.0, as shown in the following screenshot. If not, there was an error while loading requirements.txt. To debug any errors, go to the CloudWatch console and open the requirements_install_ip log in Log streams, where errors are listed. Refer to Enabling logs on the Amazon MWAA console for more details.

Set up cross-account access
You need to set up cross-account policies and roles between Account A and Account B to access the S3 buckets to load and unload data. Complete the following steps:
- In Account A, configure the bucket policy for bucket
sample-inp-bucket-etl-<username>to grant permissions to the AWS Glue and Amazon MWAA roles in Account B for objects in bucketsample-inp-bucket-etl-<username>: - Similarly, configure the bucket policy for bucket
sample-opt-bucket-etl-<username>to grant permissions to Amazon MWAA roles in Account B to put objects in this bucket: - In Account A, create an IAM policy called
policy_for_roleA, which allows necessary Amazon S3 actions on the output bucket: - Create a new IAM role called
RoleAwith Account B as the trusted entity role and add this policy to the role. This allows Account B to assume RoleA to perform necessary Amazon S3 actions on the output bucket. - In Account B, create an IAM policy called
s3-cross-account-accesswith permission to access objects in the bucketsample-inp-bucket-etl-<username>, which is in Account A. - Add this policy to the AWS Glue role and Amazon MWAA role:
- In Account B, create the IAM policy
policy_for_roleBspecifying Account A as a trusted entity. The following is the trust policy to assumeRoleAin Account A: - Create a new IAM role called
RoleBwith Amazon Redshift as the trusted entity type and add this policy to the role. This allowsRoleBto assumeRoleAin Account A and also to be assumable by Amazon Redshift. - Attach
RoleBto the Redshift Serverless namespace, so Amazon Redshift can write objects to the S3 output bucket in Account A. - Attach the policy
policy_for_roleBto the Amazon MWAA role, which allows Amazon MWAA to access the output bucket in Account A.

Refer to How do I provide cross-account access to objects that are in Amazon S3 buckets? for more details on setting up cross-account access to objects in Amazon S3 from AWS Glue and Amazon MWAA. Refer to How do I COPY or UNLOAD data from Amazon Redshift to an Amazon S3 bucket in another account? for more details on setting up roles to unload data from Amazon Redshift to Amazon S3 from Amazon MWAA.
Set up VPC peering between the Amazon MWAA and Amazon Redshift VPCs
Because Amazon MWAA and Amazon Redshift are in two separate VPCs, you need to set up VPC peering between them. You must add a route to the route tables associated with the subnets for both services. Refer to Work with VPC peering connections for details on VPC peering.

Make sure that CIDR range of the Amazon MWAA VPC is allowed in the Redshift security group and the CIDR range of the Amazon Redshift VPC is allowed in the Amazon MWAA security group, as shown in the following screenshot.


If any of the preceding steps are configured incorrectly, you are likely to encounter a “Connection Timeout” error in the DAG run.
Configure the Amazon MWAA connection with Secrets Manager
When the Amazon MWAA pipeline is configured to use Secrets Manager, it will first look for connections and variables in an alternate backend (like Secrets Manager). If the alternate backend contains the needed value, it is returned. Otherwise, it will check the metadata database for the value and return that instead. For more details, refer to Configuring an Apache Airflow connection using an AWS Secrets Manager secret.
Complete the following steps:
- Configure a VPC endpoint to link Amazon MWAA and Secrets Manager (
com.amazonaws.us-east-1.secretsmanager).
This allows Amazon MWAA to access credentials stored in Secrets Manager.

- To provide Amazon MWAA with permission to access Secrets Manager secret keys, add the policy called
SecretsManagerReadWriteto the IAM role of the environment. - To create the Secrets Manager backend as an Apache Airflow configuration option, go to the Airflow configuration options, add the following key-value pairs, and save your settings.
This configures Airflow to look for connection strings and variables at the airflow/connections/* and airflow/variables/* paths:

- To generate an Airflow connection URI string, go to AWS CloudShell and enter into a Python shell.
- Run the following code to generate the connection URI string:
The connection string should be generated as follows:
- Add the connection in Secrets Manager using the following command in the AWS Command Line Interface (AWS CLI).
This can also be done from the Secrets Manager console. This will be added in Secrets Manager as plaintext.
Use the connection airflow/connections/secrets_redshift_connection in the DAG. When the DAG is run, it will look for this connection and retrieve the secrets from Secrets Manager. In case of RedshiftDataOperator, pass the secret_arn as a parameter instead of connection name.
You can also add secrets using the Secrets Manager console as key-value pairs.
- Add another secret in Secrets Manager in and save it as
airflow/connections/redshift_conn_test.

Create an Airflow connection through the metadata database
You can also create connections in the UI. In this case, the connection details will be stored in an Airflow metadata database. If the Amazon MWAA environment is not configured to use the Secrets Manager backend, it will check the metadata database for the value and return that. You can create an Airflow connection using the UI, AWS CLI, or API. In this section, we show how to create a connection using the Airflow UI.
- For Connection Id, enter a name for the connection.
- For Connection Type, choose Amazon Redshift.
- For Host, enter the Redshift endpoint (without port and database) for Redshift Serverless.
- For Database, enter
dev. - For User, enter your admin user name.
- For Password, enter your password.
- For Port, use port 5439.
- For Extra, set the
regionandtimeoutparameters. - Test the connection, then save your settings.

Create and run a DAG
In this section, we describe how to create a DAG using various components. After you create and run the DAG, you can verify the results by querying Redshift tables and checking the target S3 buckets.
Create a DAG
In Airflow, data pipelines are defined in Python code as DAGs. We create a DAG that consists of various operators, sensors, connections, tasks, and rules:
- The DAG starts with looking for source files in the S3 bucket
sample-inp-bucket-etl-<username>under Account A for the current day usingS3KeySensor. S3KeySensor is used to wait for one or multiple keys to be present in an S3 bucket.- For example, our S3 bucket is partitioned as
s3://bucket/products/YYYY/MM/DD/, so our sensor should check for folders with the current date. We derived the current date in the DAG and passed this toS3KeySensor, which looks for any new files in the current day folder. - We also set
wildcard_matchasTrue, which enables searches onbucket_keyto be interpreted as a Unix wildcard pattern. Set themodetorescheduleso that the sensor task frees the worker slot when the criteria is not met and it’s rescheduled at a later time. As a best practice, use this mode whenpoke_intervalis more than 1 minute to prevent too much load on a scheduler.
- For example, our S3 bucket is partitioned as
- After the file is available in the S3 bucket, the AWS Glue crawler runs using
GlueCrawlerOperatorto crawl the S3 source bucketsample-inp-bucket-etl-<username>under Account A and updates the table metadata under theproducts_dbdatabase in the Data Catalog. The crawler uses the AWS Glue role and Data Catalog database that were created in the previous steps. - The DAG uses
GlueCrawlerSensorto wait for the crawler to complete. - When the crawler job is complete,
GlueJobOperatoris used to run the AWS Glue job. The AWS Glue script name (along with location) and is passed to the operator along with the AWS Glue IAM role. Other parameters likeGlueVersion,NumberofWorkers, andWorkerTypeare passed using thecreate_job_kwargsparameter. - The DAG uses
GlueJobSensorto wait for the AWS Glue job to complete. When it’s complete, the Redshift staging tableproductswill be loaded with data from the S3 file. - You can connect to Amazon Redshift from Airflow using three different operators:
PythonOperator.SQLExecuteQueryOperator, which uses a PostgreSQL connection andredshift_defaultas the default connection.RedshiftDataOperator, which uses the Redshift Data API andaws_defaultas the default connection.
In our DAG, we use SQLExecuteQueryOperator and RedshiftDataOperator to show how to use these operators. The Redshift stored procedures are run RedshiftDataOperator. The DAG also runs SQL commands in Amazon Redshift to delete the data from the staging table using SQLExecuteQueryOperator.
Because we configured our Amazon MWAA environment to look for connections in Secrets Manager, when the DAG runs, it retrieves the Redshift connection details like user name, password, host, port, and Region from Secrets Manager. If the connection is not found in Secrets Manager, the values are retrieved from the default connections.
In SQLExecuteQueryOperator, we pass the connection name that we created in Secrets Manager. It looks for airflow/connections/secrets_redshift_connection and retrieves the secrets from Secrets Manager. If Secrets Manager is not set up, the connection created manually (for example, redshift-conn-id) can be passed.
In RedshiftDataOperator, we pass the secret_arn of the airflow/connections/redshift_conn_test connection created in Secrets Manager as a parameter.
- As final task,
RedshiftToS3Operatoris used to unload data from the Redshift table to an S3 bucketsample-opt-bucket-etlin Account B.airflow/connections/redshift_conn_testfrom Secrets Manager is used for unloading the data. TriggerRuleis set toALL_DONE, which enables the next step to run after all upstream tasks are complete.- The dependency of tasks is defined using the
chain()function, which allows for parallel runs of tasks if needed. In our case, we want all tasks to run in sequence.
The following is the complete DAG code. The dag_id should match the DAG script name, otherwise it won’t be synced into the Airflow UI.
Verify the DAG run
After you create the DAG file (replace the variables in the DAG script) and upload it to the s3://sample-airflow-instance/dags folder, it will be automatically synced with the Airflow UI. All DAGs appear on the DAGs tab. Toggle the ON option to make the DAG runnable. Because our DAG is set to schedule="@once", you need to manually run the job by choosing the run icon under Actions. When the DAG is complete, the status is updated in green, as shown in the following screenshot.

In the Links section, there are options to view the code, graph, grid, log, and more. Choose Graph to visualize the DAG in a graph format. As shown in the following screenshot, each color of the node denotes a specific operator, and the color of the node outline denotes a specific status.

Verify the results
On the Amazon Redshift console, navigate to the Query Editor v2 and select the data in the products_f table. The table should be loaded and have the same number of records as S3 files.

On the Amazon S3 console, navigate to the S3 bucket s3://sample-opt-bucket-etl in Account B. The product_f files should be created under the folder structure s3://sample-opt-bucket-etl/products/YYYY/MM/DD/.

Clean up
Clean up the resources created as part of this post to avoid incurring ongoing charges:
- Delete the CloudFormation stacks and S3 bucket that you created as prerequisites.
- Delete the VPCs and VPC peering connections, cross-account policies and roles, and secrets in Secrets Manager.
Conclusion
With Amazon MWAA, you can build complex workflows using Airflow and Python without managing clusters, nodes, or any other operational overhead typically associated with deploying and scaling Airflow in production. In this post, we showed how Amazon MWAA provides an automated way to ingest, transform, analyze, and distribute data between different accounts and services within AWS. For more examples of other AWS operators, refer to the following GitHub repository; we encourage you to learn more by trying out some of these examples.
About the Authors

Radhika Jakkula is a Big Data Prototyping Solutions Architect at AWS. She helps customers build prototypes using AWS analytics services and purpose-built databases. She is a specialist in assessing wide range of requirements and applying relevant AWS services, big data tools, and frameworks to create a robust architecture.
 Sidhanth Muralidhar is a Principal Technical Account Manager at AWS. He works with large enterprise customers who run their workloads on AWS. He is passionate about working with customers and helping them architect workloads for costs, reliability, performance, and operational excellence at scale in their cloud journey. He has a keen interest in data analytics as well.
Sidhanth Muralidhar is a Principal Technical Account Manager at AWS. He works with large enterprise customers who run their workloads on AWS. He is passionate about working with customers and helping them architect workloads for costs, reliability, performance, and operational excellence at scale in their cloud journey. He has a keen interest in data analytics as well.





















 Takeshi Nakatani is a Principal Big Data Consultant on the Professional Services team in Tokyo. He has 26 years of experience in the IT industry, with expertise in architecting data infrastructure. On his days off, he can be a rock drummer or a motorcyclist.
Takeshi Nakatani is a Principal Big Data Consultant on the Professional Services team in Tokyo. He has 26 years of experience in the IT industry, with expertise in architecting data infrastructure. On his days off, he can be a rock drummer or a motorcyclist. Noritaka Sekiyama is a Principal Big Data Architect on the AWS Glue team. He is responsible for building software artifacts to help customers. In his spare time, he enjoys cycling with his road bike.
Noritaka Sekiyama is a Principal Big Data Architect on the AWS Glue team. He is responsible for building software artifacts to help customers. In his spare time, he enjoys cycling with his road bike.

 Atul Khare is a Director of Engineering at Salesforce Security, where he spearheads the Security Log Platform and Data Lakehouse initiatives. He supports diverse security customers by building robust big data ETL pipeline that is elastic, resilient, and easy to use, providing uniform & consistent security datasets for threat detection and response operations, AI, forensic analysis, analytics, and compliance needs across all Salesforce clouds. Beyond his professional endeavors, Atul enjoys performing music with his band to raise funds for local charities.
Atul Khare is a Director of Engineering at Salesforce Security, where he spearheads the Security Log Platform and Data Lakehouse initiatives. He supports diverse security customers by building robust big data ETL pipeline that is elastic, resilient, and easy to use, providing uniform & consistent security datasets for threat detection and response operations, AI, forensic analysis, analytics, and compliance needs across all Salesforce clouds. Beyond his professional endeavors, Atul enjoys performing music with his band to raise funds for local charities. Bhupender Panwar is a Big Data Architect at Salesforce and seasoned advocate for big data and cloud computing. His background encompasses the development of data-intensive applications and pipelines, solving intricate architectural and scalability challenges, and extracting valuable insights from extensive datasets within the technology industry. Outside of his big data work, Bhupender loves to hike, bike, enjoy travel and is a great foodie.
Bhupender Panwar is a Big Data Architect at Salesforce and seasoned advocate for big data and cloud computing. His background encompasses the development of data-intensive applications and pipelines, solving intricate architectural and scalability challenges, and extracting valuable insights from extensive datasets within the technology industry. Outside of his big data work, Bhupender loves to hike, bike, enjoy travel and is a great foodie. Avijit Goswami is a Principal Solutions Architect at AWS specialized in data and analytics. He supports AWS strategic customers in building high-performing, secure, and scalable data lake solutions on AWS using AWS managed services and open-source solutions. Outside of his work, Avijit likes to travel, hike in the San Francisco Bay Area trails, watch sports, and listen to music.
Avijit Goswami is a Principal Solutions Architect at AWS specialized in data and analytics. He supports AWS strategic customers in building high-performing, secure, and scalable data lake solutions on AWS using AWS managed services and open-source solutions. Outside of his work, Avijit likes to travel, hike in the San Francisco Bay Area trails, watch sports, and listen to music. Vikas Panghal is the Principal Product Manager leading the product management team for Amazon SNS and Amazon SQS. He has deep expertise in event-driven and messaging applications and brings a wealth of knowledge and experience to his role, shaping the future of messaging services. He is passionate about helping customers build highly scalable, fault-tolerant, and loosely coupled systems. Outside of work, he enjoys spending time with his family outdoors, playing chess, and running.
Vikas Panghal is the Principal Product Manager leading the product management team for Amazon SNS and Amazon SQS. He has deep expertise in event-driven and messaging applications and brings a wealth of knowledge and experience to his role, shaping the future of messaging services. He is passionate about helping customers build highly scalable, fault-tolerant, and loosely coupled systems. Outside of work, he enjoys spending time with his family outdoors, playing chess, and running.

















 Utkarsh Mittal is a Senior Technical Product Manager for Amazon DataZone at AWS. He is passionate about building innovative products that simplify customers’ end-to-end analytics journeys. Outside of the tech world, Utkarsh loves to play music, with drums being his latest endeavor.
Utkarsh Mittal is a Senior Technical Product Manager for Amazon DataZone at AWS. He is passionate about building innovative products that simplify customers’ end-to-end analytics journeys. Outside of the tech world, Utkarsh loves to play music, with drums being his latest endeavor. Praveen Kumar is a Principal Analytics Solution Architect at AWS with expertise in designing, building, and implementing modern data and analytics platforms using cloud-centered services. His areas of interests are serverless technology, modern cloud data warehouses, streaming, and generative AI applications.
Praveen Kumar is a Principal Analytics Solution Architect at AWS with expertise in designing, building, and implementing modern data and analytics platforms using cloud-centered services. His areas of interests are serverless technology, modern cloud data warehouses, streaming, and generative AI applications. Paul Villena is a Senior Analytics Solutions Architect in AWS with expertise in building modern data and analytics solutions to drive business value. He works with customers to help them harness the power of the cloud. His areas of interests are infrastructure as code, serverless technologies, and coding in Python
Paul Villena is a Senior Analytics Solutions Architect in AWS with expertise in building modern data and analytics solutions to drive business value. He works with customers to help them harness the power of the cloud. His areas of interests are infrastructure as code, serverless technologies, and coding in Python
























 Andrea Filippo is a Partner Solutions Architect at AWS supporting Public Sector partners and customers in Italy. He focuses on modern data architectures and helping customers accelerate their cloud journey with serverless technologies.
Andrea Filippo is a Partner Solutions Architect at AWS supporting Public Sector partners and customers in Italy. He focuses on modern data architectures and helping customers accelerate their cloud journey with serverless technologies. Emanuele is a Solutions Architect at AWS, based in Italy, after living and working for more than 5 years in Spain. He enjoys helping large companies with the adoption of cloud technologies, and his area of expertise is mainly focused on Data Analytics and Data Management. Outside of work, he enjoys traveling and collecting action figures.
Emanuele is a Solutions Architect at AWS, based in Italy, after living and working for more than 5 years in Spain. He enjoys helping large companies with the adoption of cloud technologies, and his area of expertise is mainly focused on Data Analytics and Data Management. Outside of work, he enjoys traveling and collecting action figures. Varsha Velagapudi is a Senior Technical Product Manager with Amazon DataZone at AWS. She focuses on improving data discovery and curation required for data analytics. She is passionate about simplifying customers’ AI/ML and analytics journey to help them succeed in their day-to-day tasks. Outside of work, she enjoys nature and outdoor activities, reading, and traveling.
Varsha Velagapudi is a Senior Technical Product Manager with Amazon DataZone at AWS. She focuses on improving data discovery and curation required for data analytics. She is passionate about simplifying customers’ AI/ML and analytics journey to help them succeed in their day-to-day tasks. Outside of work, she enjoys nature and outdoor activities, reading, and traveling.

 Andries Engelbrecht is a Principal Partner Solutions Architect at Snowflake and works with strategic partners. He is actively engaged with strategic partners like AWS supporting product and service integrations as well as the development of joint solutions with partners. Andries has over 20 years of experience in the field of data and analytics.
Andries Engelbrecht is a Principal Partner Solutions Architect at Snowflake and works with strategic partners. He is actively engaged with strategic partners like AWS supporting product and service integrations as well as the development of joint solutions with partners. Andries has over 20 years of experience in the field of data and analytics. Deenbandhu Prasad is a Senior Analytics Specialist at AWS, specializing in big data services. He is passionate about helping customers build modern data architectures on the AWS Cloud. He has helped customers of all sizes implement data management, data warehouse, and data lake solutions.
Deenbandhu Prasad is a Senior Analytics Specialist at AWS, specializing in big data services. He is passionate about helping customers build modern data architectures on the AWS Cloud. He has helped customers of all sizes implement data management, data warehouse, and data lake solutions. Brian Dolan joined Amazon as a Military Relations Manager in 2012 after his first career as a Naval Aviator. In 2014, Brian joined Amazon Web Services, where he helped Canadian customers from startups to enterprises explore the AWS Cloud. Most recently, Brian was a member of the Non-Relational Business Development team as a Go-To-Market Specialist for Amazon DynamoDB and Amazon Keyspaces before joining the Analytics Worldwide Specialist Organization in 2022 as a Go-To-Market Specialist for AWS Glue.
Brian Dolan joined Amazon as a Military Relations Manager in 2012 after his first career as a Naval Aviator. In 2014, Brian joined Amazon Web Services, where he helped Canadian customers from startups to enterprises explore the AWS Cloud. Most recently, Brian was a member of the Non-Relational Business Development team as a Go-To-Market Specialist for Amazon DynamoDB and Amazon Keyspaces before joining the Analytics Worldwide Specialist Organization in 2022 as a Go-To-Market Specialist for AWS Glue. Nidhi Gupta is a Sr. Partner Solution Architect at AWS. She spends her days working with customers and partners, solving architectural challenges. She is passionate about data integration and orchestration, serverless and big data processing, and machine learning. Nidhi has extensive experience leading the architecture design and production release and deployments for data workloads.
Nidhi Gupta is a Sr. Partner Solution Architect at AWS. She spends her days working with customers and partners, solving architectural challenges. She is passionate about data integration and orchestration, serverless and big data processing, and machine learning. Nidhi has extensive experience leading the architecture design and production release and deployments for data workloads. Scott Teal is a Product Marketing Lead at Snowflake and focuses on data lakes, storage, and governance.
Scott Teal is a Product Marketing Lead at Snowflake and focuses on data lakes, storage, and governance.












 Noritaka Sekiyama is a Principal Big Data Architect on the AWS Glue team. He is responsible for building software artifacts to help customers. In his spare time, he enjoys cycling with his new road bike.
Noritaka Sekiyama is a Principal Big Data Architect on the AWS Glue team. He is responsible for building software artifacts to help customers. In his spare time, he enjoys cycling with his new road bike. Chuhan Liu is a Software Development Engineer on the AWS Glue team. He is passionate about building scalable distributed systems for big data processing, analytics, and management. In his spare time, he enjoys playing tennis.
Chuhan Liu is a Software Development Engineer on the AWS Glue team. He is passionate about building scalable distributed systems for big data processing, analytics, and management. In his spare time, he enjoys playing tennis. XiaoRun Yu is a Software Development Engineer on the AWS Glue team. He is working on building new features for AWS Glue to help customers. Outside of work, Xiaorun enjoys exploring new places in the Bay Area.
XiaoRun Yu is a Software Development Engineer on the AWS Glue team. He is working on building new features for AWS Glue to help customers. Outside of work, Xiaorun enjoys exploring new places in the Bay Area. Sean Ma is a Principal Product Manager on the AWS Glue team. He has a track record of more than 18 years innovating and delivering enterprise products that unlock the power of data for users. Outside of work, Sean enjoys scuba diving and college football.
Sean Ma is a Principal Product Manager on the AWS Glue team. He has a track record of more than 18 years innovating and delivering enterprise products that unlock the power of data for users. Outside of work, Sean enjoys scuba diving and college football. Mohit Saxena is a Senior Software Development Manager on the AWS Glue team. His team focuses on building distributed systems to enable customers with interactive and simple to use interfaces to efficiently manage and transform petabytes of data seamlessly across data lakes on Amazon S3, databases and data-warehouses on cloud.
Mohit Saxena is a Senior Software Development Manager on the AWS Glue team. His team focuses on building distributed systems to enable customers with interactive and simple to use interfaces to efficiently manage and transform petabytes of data seamlessly across data lakes on Amazon S3, databases and data-warehouses on cloud.







 Sushanth Kothapally is a Solutions Architect at Amazon Web Services supporting Automotive and Manufacturing customers. He is passionate about designing technology solutions to meet business goals and has keen interest in serverless and event-driven architectures.
Sushanth Kothapally is a Solutions Architect at Amazon Web Services supporting Automotive and Manufacturing customers. He is passionate about designing technology solutions to meet business goals and has keen interest in serverless and event-driven architectures. Senthil Kamala Rathinam is a Solutions Architect at Amazon Web Services specializing in Data and Analytics. He is passionate about helping customers to design and build modern data platforms. In his free time, Senthil loves to spend time with his family and play badminton.
Senthil Kamala Rathinam is a Solutions Architect at Amazon Web Services specializing in Data and Analytics. He is passionate about helping customers to design and build modern data platforms. In his free time, Senthil loves to spend time with his family and play badminton.



















 Alan Peaty is a Senior Partner Solutions Architect at AWS. Alan helps Global Systems Integrators (GSIs) and Global Independent Software Vendors (GISVs) solve complex customer challenges using AWS services. Prior to joining AWS, Alan worked as an architect at systems integrators to translate business requirements into technical solutions. Outside of work, Alan is an IoT enthusiast and a keen runner who loves to hit the muddy trails of the English countryside.
Alan Peaty is a Senior Partner Solutions Architect at AWS. Alan helps Global Systems Integrators (GSIs) and Global Independent Software Vendors (GISVs) solve complex customer challenges using AWS services. Prior to joining AWS, Alan worked as an architect at systems integrators to translate business requirements into technical solutions. Outside of work, Alan is an IoT enthusiast and a keen runner who loves to hit the muddy trails of the English countryside. Parag Srivastava is a Solutions Architect at AWS, helping enterprise customers with successful cloud adoption and migration. During his professional career, he has been extensively involved in complex digital transformation projects. He is also passionate about building innovative solutions around geospatial aspects of addresses.
Parag Srivastava is a Solutions Architect at AWS, helping enterprise customers with successful cloud adoption and migration. During his professional career, he has been extensively involved in complex digital transformation projects. He is also passionate about building innovative solutions around geospatial aspects of addresses.















































 Satya Adimula is a Senior Data Architect at AWS based in Boston. With over two decades of experience in data and analytics, Satya helps organizations derive business insights from their data at scale.
Satya Adimula is a Senior Data Architect at AWS based in Boston. With over two decades of experience in data and analytics, Satya helps organizations derive business insights from their data at scale.















 Toney Thomas is a Data Architect and Data Engineering Lead at Bluestone, renowned for his role in envisioning and coining the company’s pioneering data strategy. With a strategic focus on harnessing the power of advanced technology to tackle intricate business challenges, Toney leads a dynamic team of Data Engineers, Reporting Engineers, Quality Assurance specialists, and Business Analysts at Bluestone. His leadership extends to driving the implementation of robust data governance frameworks across diverse organizational units. Under his guidance, Bluestone has achieved remarkable success, including the deployment of innovative platforms such as a fully governed data mesh business data system with embedded data quality mechanisms, aligning seamlessly with the organization’s commitment to data democratization and excellence.
Toney Thomas is a Data Architect and Data Engineering Lead at Bluestone, renowned for his role in envisioning and coining the company’s pioneering data strategy. With a strategic focus on harnessing the power of advanced technology to tackle intricate business challenges, Toney leads a dynamic team of Data Engineers, Reporting Engineers, Quality Assurance specialists, and Business Analysts at Bluestone. His leadership extends to driving the implementation of robust data governance frameworks across diverse organizational units. Under his guidance, Bluestone has achieved remarkable success, including the deployment of innovative platforms such as a fully governed data mesh business data system with embedded data quality mechanisms, aligning seamlessly with the organization’s commitment to data democratization and excellence. Ben Vengerovsky is a Data Platform Product Manager at Bluestone. He is passionate about using cloud technology to revolutionize the company’s data infrastructure. With a background in mortgage lending and a deep understanding of AWS services, Ben specializes in designing scalable and efficient data solutions that drive business growth and enhance customer experiences. He thrives on collaborating with cross-functional teams to translate business requirements into innovative technical solutions that empower data-driven decision-making.
Ben Vengerovsky is a Data Platform Product Manager at Bluestone. He is passionate about using cloud technology to revolutionize the company’s data infrastructure. With a background in mortgage lending and a deep understanding of AWS services, Ben specializes in designing scalable and efficient data solutions that drive business growth and enhance customer experiences. He thrives on collaborating with cross-functional teams to translate business requirements into innovative technical solutions that empower data-driven decision-making. Rada Stanic is a Chief Technologist at Amazon Web Services, where she helps ANZ customers across different segments solve their business problems using AWS Cloud technologies. Her special areas of interest are data analytics, machine learning/AI, and application modernization.
Rada Stanic is a Chief Technologist at Amazon Web Services, where she helps ANZ customers across different segments solve their business problems using AWS Cloud technologies. Her special areas of interest are data analytics, machine learning/AI, and application modernization.






 Michael Greenshtein is an Analytics Specialist Solutions Architect for the Public Sector.
Michael Greenshtein is an Analytics Specialist Solutions Architect for the Public Sector. Gonzalo Herreros is a Senior Big Data Architect on the AWS Glue team.
Gonzalo Herreros is a Senior Big Data Architect on the AWS Glue team.








 Xiaoxi Liu is a Software Development Engineer on the AWS Glue team. Her passion is building scalable distributed systems for efficiently managing big data on the cloud, and her concentrations are distributed system, big data, and cloud computing.
Xiaoxi Liu is a Software Development Engineer on the AWS Glue team. Her passion is building scalable distributed systems for efficiently managing big data on the cloud, and her concentrations are distributed system, big data, and cloud computing. Akira Ajisaka is a Senior Software Development Engineer on the AWS Glue team. He likes open source software and distributed systems. In his spare time, he enjoys playing arcade games.
Akira Ajisaka is a Senior Software Development Engineer on the AWS Glue team. He likes open source software and distributed systems. In his spare time, he enjoys playing arcade games. Shenoda Guirguis is a Senior Software Development Engineer on the AWS Glue team. His passion is in building scalable and distributed data infrastructure and processing systems. When he gets a chance, Shenoda enjoys reading and playing soccer.
Shenoda Guirguis is a Senior Software Development Engineer on the AWS Glue team. His passion is in building scalable and distributed data infrastructure and processing systems. When he gets a chance, Shenoda enjoys reading and playing soccer.













 Durga Prasad is a Sr Lead Consultant enabling customers build their Data Analytics solutions on AWS. He is a coffee lover and enjoys playing badminton.
Durga Prasad is a Sr Lead Consultant enabling customers build their Data Analytics solutions on AWS. He is a coffee lover and enjoys playing badminton. Murali Reddy is a Lead Consultant at Amazon Web Services (AWS), helping customers build and implement data analytics solution. When he’s not working, Murali is an avid bike rider and loves exploring new places.
Murali Reddy is a Lead Consultant at Amazon Web Services (AWS), helping customers build and implement data analytics solution. When he’s not working, Murali is an avid bike rider and loves exploring new places.




















 Michael Hamilton is a Sr Analytics Solutions Architect focusing on helping enterprise customers modernize and simplify their analytics workloads on AWS. He enjoys mountain biking and spending time with his wife and three children when not working.
Michael Hamilton is a Sr Analytics Solutions Architect focusing on helping enterprise customers modernize and simplify their analytics workloads on AWS. He enjoys mountain biking and spending time with his wife and three children when not working. Daniel Rozo is a Senior Solutions Architect with AWS supporting customers in the Netherlands. His passion is engineering simple data and analytics solutions and helping customers move to modern data architectures. Outside of work, he enjoys playing tennis and biking.
Daniel Rozo is a Senior Solutions Architect with AWS supporting customers in the Netherlands. His passion is engineering simple data and analytics solutions and helping customers move to modern data architectures. Outside of work, he enjoys playing tennis and biking.