Post Syndicated from Amir Shenavandeh original https://aws.amazon.com/blogs/big-data/get-a-quick-start-with-apache-hudi-apache-iceberg-and-delta-lake-with-amazon-emr-on-eks/
A data lake is a centralized repository that allows you to store all your structured and unstructured data at any scale. You can keep your data as is in your object store or file-based storage without having to first structure the data. Additionally, you can run different types of analytics against your loosely formatted data lake—from dashboards and visualizations to big data processing, real-time analytics, and machine learning (ML) to guide better decisions. Due to the flexibility and cost effectiveness that a data lake offers, it’s very popular with customers who are looking to implement data analytics and AI/ML use cases.
Due to the immutable nature of the underlying storage in the cloud, one of the challenges in data processing is updating or deleting a subset of identified records from a data lake. Another challenge is making concurrent changes to the data lake. Implementing these tasks is time consuming and costly.
In this post, we explore three open-source transactional file formats: Apache Hudi, Apache Iceberg, and Delta Lake to help us to overcome these data lake challenges. We focus on how to get started with these data storage frameworks via real-world use case. As an example, we demonstrate how to handle incremental data change in a data lake by implementing a Slowly Changing Dimension Type 2 solution (SCD2) with Hudi, Iceberg, and Delta Lake, then deploy the applications with Amazon EMR on EKS.
ACID challenge in data lakes
In analytics, the data lake plays an important role as an immutable and agile data storage layer. Unlike traditional data warehouses or data mart implementations, we make no assumptions on the data schema in a data lake and can define whatever schemas required by our use cases. It’s up to the downstream consumption layer to make sense of that data for their own purposes.
One of the most common challenges is supporting ACID (Atomicity, Consistency, Isolation, Durability) transactions in a data lake. For example, how do we run queries that return consistent and up-to-date results while new data is continuously being ingested or existing data is being modified?
Let’s try to understand the data problem with a real-world scenario. Assume we centralize customer contact datasets from multiple sources to an Amazon Simple Storage Service (Amazon S3)-backed data lake, and we want to keep all the historical records for analysis and reporting. We face the following challenges:
- We keep creating append-only files in Amazon S3 to track the contact data changes (insert, update, delete) in near-real time.
- Consistency and atomicity aren’t guaranteed because we just dump data files from multiple sources without knowing whether the entire operation is successful or not.
- We don’t have an isolation guarantee whenever multiple workloads are simultaneously reading and writing to the same target contact table.
- We track every single activity at source, including duplicates caused by the retry mechanism and accidental data changes that are then reverted. This leads to the creation of a large volume of append-only files. The performance of extract, transform, and load (ETL) jobs decreases as all the data files are read each time.
- We have to shorten the file retention period to reduce the data scan and read performance.
In this post, we walk through a simple SCD2 ETL example designed for solving the ACID transaction problem with the help of Hudi, Iceberg, and Delta Lake. We also show how to deploy the ACID solution with EMR on EKS and query the results by Amazon Athena.
Custom library dependencies with EMR on EKS
By default, Hudi and Iceberg are supported by Amazon EMR as out-of-the-box features. For this demonstration, we use EMR on EKS release 6.8.0, which contains Apache Iceberg 0.14.0-amzn-0 and Apache Hudi 0.11.1-amzn-0. To find out the latest and past versions that Amazon EMR supports, check out the Hudi release history and the Iceberg release history tables. The runtime binary files of these frameworks can be found in the Spark’s class path location within each EMR on EKS image. See Amazon EMR on EKS release versions for the list of supported versions and applications.
As of this writing, Amazon EMR does not include Delta Lake by default. There are two ways to make it available in EMR on EKS:
- At the application level – You install Delta libraries by setting a Spark configuration spark.jars or
--jarscommand-line argument in your submission script. The JAR files will be downloaded and distributed to each Spark Executor and Driver pod when starting a job. - At the Docker container level – You can customize an EMR on EKS image by packaging Delta dependencies into a single Docker container that promotes portability and simplifies dependency management for each workload
Other custom library dependencies can be managed the same way as for Delta Lake—passing a comma-separated list of JAR files in the Spark configuration at job submission, or packaging all the required libraries into a Docker image.
Solution overview
The solution provides two sample CSV files as the data source: initial_contact.csv and update_contacts.csv. They were generated by a Python script with the Faker package. For more details, check out the tutorial on GitHub.
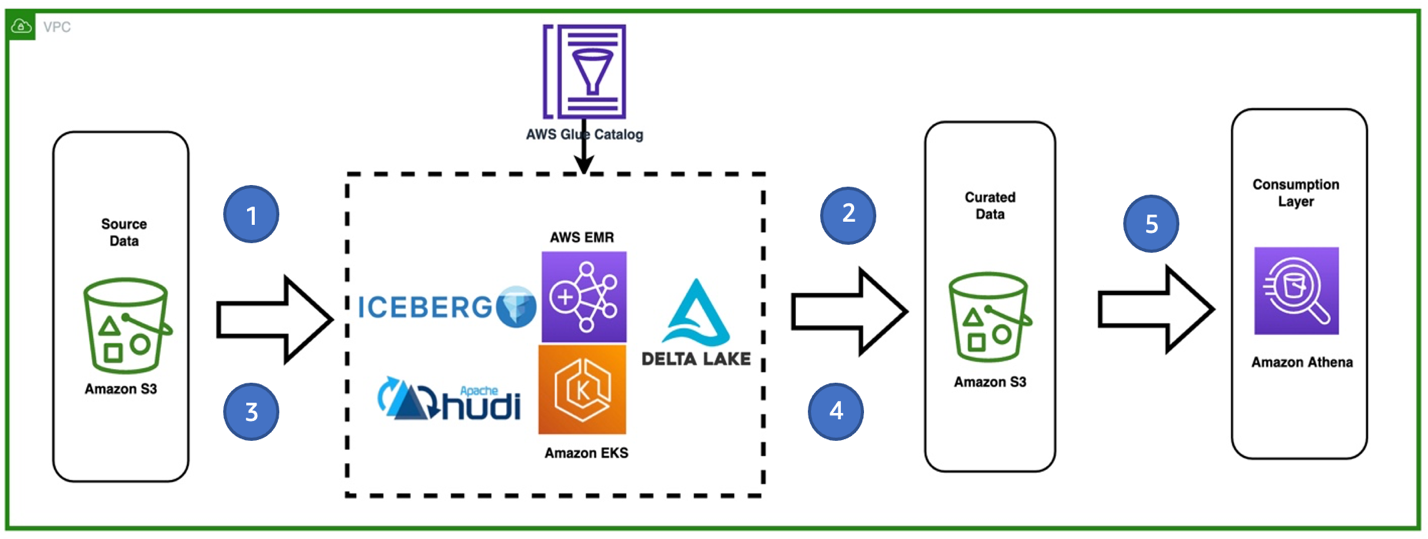
The following diagram describes a high-level architecture of the solution and different services being used.
The workflow steps are as follows:
- Ingest the first CSV file from a source S3 bucket. The data is being processed by running a Spark ETL job with EMR on EKS. The application contains either the Hudi, Iceberg, or Delta framework.
- Store the initial table in Hudi, Iceberg, or Delta file format in a target S3 bucket (curated). We use the AWS Glue Data Catalog as the hive metastore. Optionally, you can configure Amazon DynamoDB as a lock manager for the concurrency controls.
- Ingest a second CSV file that contains new records and some changes to the existing ones.
- Perform SCD2 via Hudi, Iceberg, or Delta in the Spark ETL job.
- Query the Hudi, Iceberg, or Delta table stored on the target S3 bucket in Athena
To simplify the demo, we have accommodated steps 1–4 into a single Spark application.
Prerequisites
Install the following tools:
- The AWS Command Line Interface (AWS CLI). For instructions, refer to Install the AWS CLI.
- kubectl, eksctl, and helm version 3.8.2.
For a quick start, you can use AWS CloudShell which includes the AWS CLI and kubectl already.
Clone the project
Download the sample project either to your computer or the CloudShell console:
Set up the environment
Run the following blog_provision.sh script to set up a test environment. The infrastructure deployment includes the following resources:
- A new S3 bucket to store sample data and job code.
- An Amazon Elastic Kubernetes Service (Amazon EKS) cluster (version 1.21) in a new VPC across two Availability Zones.
- An EMR virtual cluster in the same VPC, registered to the emr namespace in Amazon EKS.
- An AWS Identity and Access Management (IAM) job execution role contains DynamoDB access, because we use DynamoDB to provide concurrency controls that ensure atomic transaction with the Hudi and Iceberg tables.
Job execution role
The provisioning includes an IAM job execution role called emr-on-eks-quickstart-execution-role that allows your EMR on EKS jobs access to the required AWS services. It contains AWS Glue permissions because we use the Data Catalog as our metastore.
See the following code:
Additionally, the role contains DynamoDB permissions, because we use the service as the lock manager. It provides concurrency controls that ensure atomic transaction with our Hudi and Iceberg tables. If a DynamoDB table with the given name doesn’t exist, a new table is created with the billing mode set as pay-per-request. More details can be found in the following framework examples.
Example 1: Run Apache Hudi with EMR on EKS
The following steps provide a quick start for you to implement SCD Type 2 data processing with the Hudi framework. To learn more, refer to Build Slowly Changing Dimensions Type 2 (SCD2) with Apache Spark and Apache Hudi on Amazon EMR.
The following code snippet demonstrates the SCD type2 implementation logic. It creates Hudi tables in a default database in the Glue Data Catalog. The full version is in the script hudi_scd_script.py.
In the job script, the hudiOptions were set to use the AWS Glue Data Catalog and enable the DynamoDB-based Optimistic Concurrency Control (OCC). For more information about concurrency control and alternatives for lock providers, refer to Concurrency Control.
- Upload the job scripts to Amazon S3:
- Submit Hudi jobs with EMR on EKS to create SCD2 tables:
Hudi supports two tables types: Copy on Write (CoW) and Merge on Read (MoR). The following is the code snippet to create a CoW table. For the complete job scripts for each table type, refer to hudi_submit_cow.sh and hudi_submit_mor.sh.
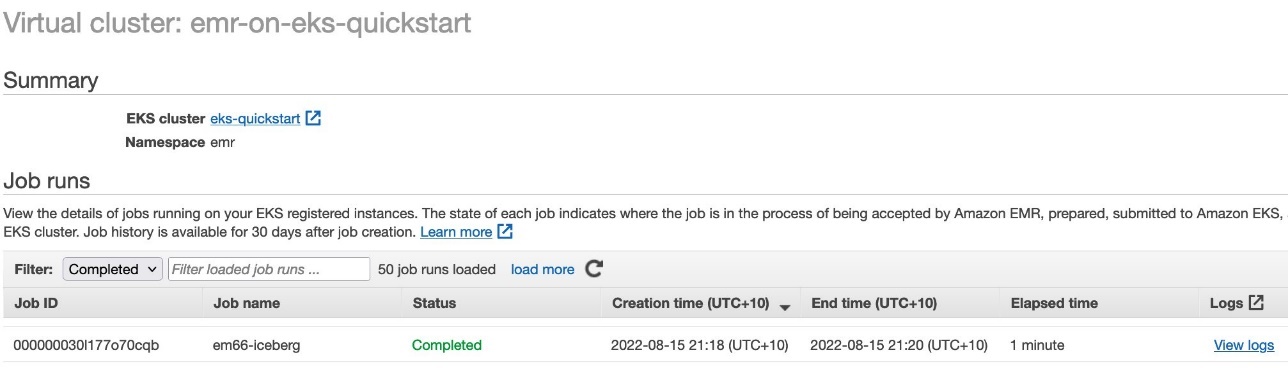
- Check the job status on the EMR virtual cluster console.

- Query the output in Athena:
Example 2: Run Apache Iceberg with EMR on EKS
Starting with Amazon EMR version 6.6.0, you can use Apache Spark 3 on EMR on EKS with the Iceberg table format. For more information on how Iceberg works in an immutable data lake, see Build a high-performance, ACID compliant, evolving data lake using Apache Iceberg on Amazon EMR.
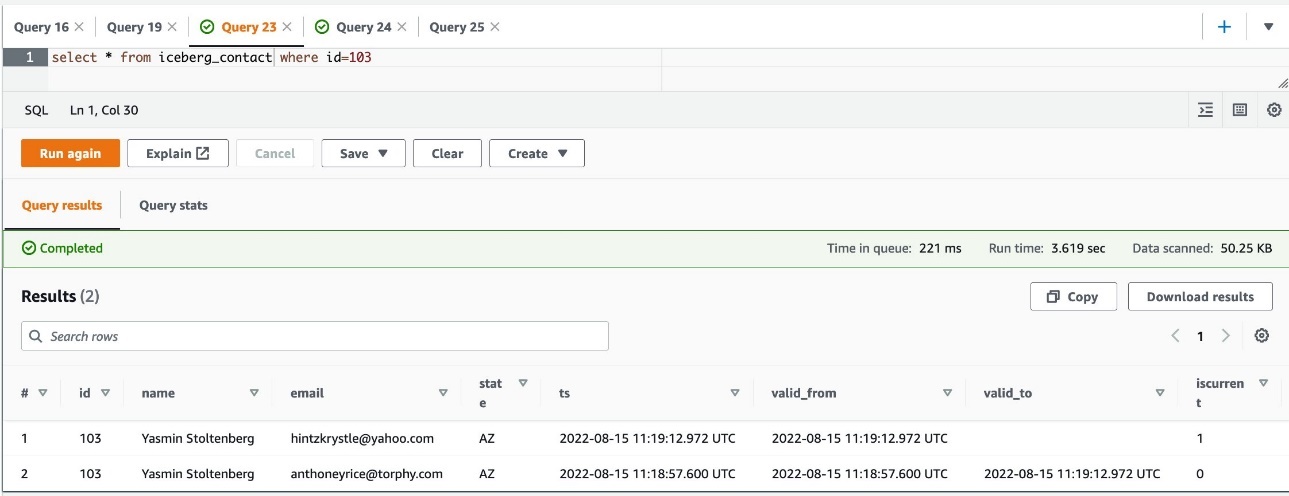
The sample job creates an Iceberg table iceberg_contact in the default database of AWS Glue. The full version is in the iceberg_scd_script.py script. The following code snippet shows the SCD2 type of MERGE operation:
As demonstrated earlier when discussing the job execution role, the role emr-on-eks-quickstart-execution-role granted access to the required DynamoDB table myIcebergLockTable, because the table is used to obtain locks on Iceberg tables, in case of multiple concurrent write operations against a single table. For more information on Iceberg’s lock manager, refer to DynamoDB Lock Manager.
- Upload the application scripts to the example S3 bucket:
- Submit the job with EMR on EKS to create an SCD2 Iceberg table:
The full version code is in the iceberg_submit.sh script. The code snippet is as follows:
- Check the job status on the EMR on EKS console.

- When the job is complete, query the table in Athena:
Example 3: Run open-source Delta Lake with EMR on EKS
Delta Lake 2.1.x is compatible with Apache Spark 3.3.x. Check out the compatibility list for other versions of Delta Lake and Spark. In this post, we use Amazon EMR release 6.8 (Spark 3.3.0) to demonstrate the SCD2 implementation in a data lake.
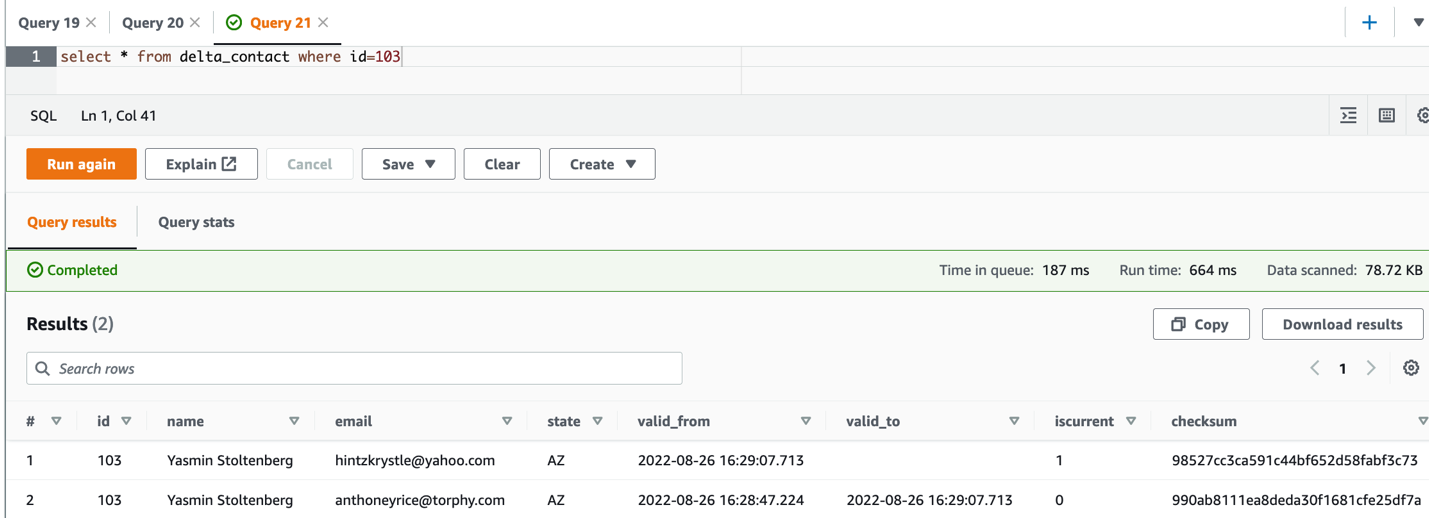
The following is the Delta code snippet to load initial dataset; the incremental load MERGE logic is highly similar to the Iceberg example. As a one-off task, there should be two tables set up on the same data:
- The Delta table delta_table_contact – Defined on the
TABLE_LOCATION at ‘s3://{S3_BUCKET_NAME}/delta/delta_contact’. The MERGE/UPSERT operation must be implemented on the Delta destination table. Athena can’t query this table directly, instead it reads from a manifest file stored in the same location, which is a text file containing a list of data files to read for querying a table. It is described as an Athena table below. - The Athena table delta_contact – Defined on the manifest location
s3://{S3_BUCKET_NAME}/delta/delta_contact/_symlink_format_manifest/. All read operations from Athena must use this table.
The full version code is in the delta_scd_script.py script. The code snippet is as follows:
The SQL statement GENERATE symlink_format_manifest FOR TABLE ... is a required step to set up the Athena for Delta Lake. Whenever the data in a Delta table is updated, you must regenerate the manifests. Therefore, we use ALTER TABLE .... SET TBLPROPERTIES(delta.compatibility.symlinkFormatManifest.enabled=true) to automate the manifest refresh as a one-off setup.
- Upload the Delta sample scripts to the S3 bucket:
- Submit the job with EMR on EKS:
The full version code is in the delta_submit.sh script. The open-source Delta JAR files must be included in the spark.jars. Alternatively, follow the instructions in How to customize Docker images and build a custom EMR on EKS image to accommodate the Delta dependencies.
The code snippet is as follows:
- Check the job status from the EMR on EKS console.

- When the job is complete, query the table in Athena:
Clean up
To avoid incurring future charges, delete the resources generated if you don’t need the solution anymore. Run the following cleanup script (change the Region if necessary):
Conclusion
Implementing an ACID-compliant data lake with EMR on EKS enables you focus more on delivering business value, instead of worrying about managing complexities and reliabilities at the data storage layer.
This post presented three different transactional storage frameworks that can meet your ACID needs. They ensure you never read partial data (Atomicity). The read/write isolation allows you to see consistent snapshots of the data, even if an update occurs at the same time (Consistency and Isolation). All the transactions are stored directly to the underlying Amazon S3-backed data lake, which is designed for 11 9’s of durability (Durability).
For more information, check out the sample GitHub repository used in this post and the EMR on EKS Workshop. They will get you started with running your familiar transactional framework with EMR on EKS. If you want dive deep into each storage format, check out the following posts:
- Build Slowly Changing Dimensions Type 2 (SCD2) with Apache Spark and Apache Hudi on Amazon EMR
- Build a high-performance, ACID compliant, evolving data lake using Apache Iceberg on Amazon EMR
- Build a high-performance, transactional data lake using Open-Source Delta Lake on Amazon EMR
About the authors
 Amir Shenavandeh is a Sr Analytics Specialist Solutions Architect and Amazon EMR subject matter expert at Amazon Web Services. He helps customers with architectural guidance and optimisation. He leverages his experience to help people bring their ideas to life, focusing on distributed processing and big data architectures.
Amir Shenavandeh is a Sr Analytics Specialist Solutions Architect and Amazon EMR subject matter expert at Amazon Web Services. He helps customers with architectural guidance and optimisation. He leverages his experience to help people bring their ideas to life, focusing on distributed processing and big data architectures.
 Melody Yang is a Senior Big Data Solutions Architect for Amazon EMR at AWS. She is an experienced analytics leader working with AWS customers to provide best practice guidance and technical advice in order to assist their success in data transformation. Her areas of interests are open-source frameworks and automation, data engineering, and DataOps.
Melody Yang is a Senior Big Data Solutions Architect for Amazon EMR at AWS. She is an experienced analytics leader working with AWS customers to provide best practice guidance and technical advice in order to assist their success in data transformation. Her areas of interests are open-source frameworks and automation, data engineering, and DataOps.
 Amit Maindola is a Data Architect focused on big data and analytics at Amazon Web Services. He helps customers in their digital transformation journey and enables them to build highly scalable, robust, and secure cloud-based analytical solutions on AWS to gain timely insights and make critical business decisions.
Amit Maindola is a Data Architect focused on big data and analytics at Amazon Web Services. He helps customers in their digital transformation journey and enables them to build highly scalable, robust, and secure cloud-based analytical solutions on AWS to gain timely insights and make critical business decisions.