Post Syndicated from Pradeep Misra original https://aws.amazon.com/blogs/big-data/use-your-corporate-identities-for-analytics-with-amazon-emr-and-aws-iam-identity-center/
To enable your workforce users for analytics with fine-grained data access controls and audit data access, you might have to create multiple AWS Identity and Access Management (IAM) roles with different data permissions and map the workforce users to one of those roles. Multiple users are often mapped to the same role where they need similar privileges to enable data access controls at the corporate user or group level and audit data access.
AWS IAM Identity Center enables centralized management of workforce user access to AWS accounts and applications using a local identity store or by connecting corporate directories via identity providers (IdPs). IAM Identity Center now supports trusted identity propagation, a streamlined experience for users who require access to data with AWS analytics services.
Amazon EMR Studio is an integrated development environment (IDE) that makes it straightforward for data scientists and data engineers to build data engineering and data science applications. With trusted identity propagation, data access management can be based on a user’s corporate identity and can be propagated seamlessly as they access data with single sign-on to build analytics applications with Amazon EMR (EMR Studio and Amazon EMR on EC2).
AWS Lake Formation allows data administrators to centrally govern, secure, and share data for analytics and machine learning (ML). With trusted identity propagation, data administrators can directly provide granular access to corporate users using their identity attributes and simplify the traceability of end-to-end data access across AWS services. Because access is managed based on a user’s corporate identity, they don’t need to use database local user credentials or assume an IAM role to access data.
In this post, we show how to bring your workforce identity to EMR Studio for analytics use cases, directly manage fine-grained permissions for the corporate users and groups using Lake Formation, and audit their data access.
Solution overview
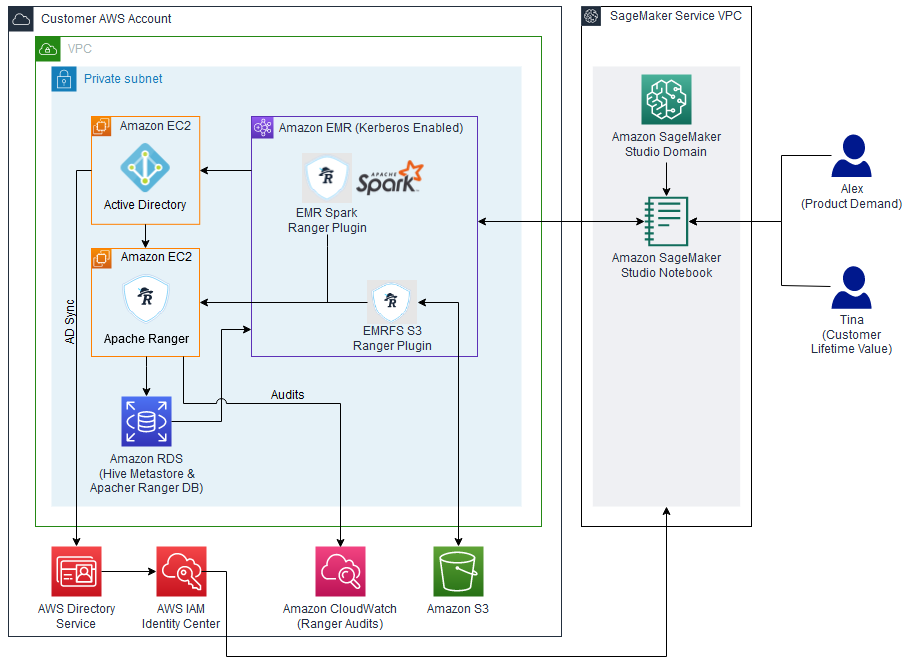
For our use case, we want to enable a data analyst user named analyst1 to use their own enterprise credentials to query data they have been granted permissions to and audit their data access. We use Okta as the IdP for this demonstration. The following diagram illustrates the solution architecture.

This architecture is based on the following components:
- Okta is responsible for maintaining the corporate user identities, related groups, and user authentication.
- IAM Identity Center connects Okta users and centrally manages their access across AWS accounts and applications.
- Lake Formation provides fine-grained access controls on data directly to corporate users using trusted identity propagation.
- EMR Studio is an IDE for users to build and run applications. It allows users to log in directly with their corporate credentials without signing in to the AWS Management Console.
- AWS Service Catalog provides a product template to create EMR clusters.
- EMR cluster is integrated with IAM Identity Center using a security configuration.
- AWS CloudTrail captures user data access activities.
The following are the high-level steps to implement the solution:
- Integrate Okta with IAM Identity Center.
- Set up Amazon EMR Studio.
- Create an IAM Identity Center enabled security configuration for EMR clusters.
- Create a Service Catalog product template to create the EMR clusters.
- Use Lake Formation to grant permissions to users to access data.
- Test the solution by accessing data with a corporate identity.
- Audit user data access.
Prerequisites
You should have the following prerequisites:
- An AWS account with access to the following AWS services:
- AWS CloudFormation
- CloudTrail
- Amazon Elastic Compute Cloud (Amazon EC2)
- Amazon Simple Storage Service (Amazon S3)
- EMR Studio
- IAM
- IAM Identity Center
- Lake Formation
- Service Catalog
- An Okta account (you can create a free developer account)
Integrate Okta with IAM Identity Center
For more information about configuring Okta with IAM Identity Center, refer to Configure SAML and SCIM with Okta and IAM Identity Center.
For this setup, we have created two users, analyst1 and engineer1, and assigned them to the corresponding Okta application. You can validate the integration is working by navigating to the Users page on the IAM Identity Center console, as shown in the following screenshot. Both enterprise users from Okta are provisioned in IAM Identity Center.

The following exact users will not be listed in your account. You can either create similar users or use an existing user.

Each provisioned user in IAM Identity Center has a unique user ID. This ID does not originate from Okta; it’s created in IAM Identity Center to uniquely identify this user. With trusted identity propagation, this user ID will be propagated across services and also used for traceability purposes in CloudTrail. The following screenshot shows the IAM Identity Center user matching the provisioned Okta user analyst1.

Choose the link under AWS access portal URL and log in with the analyst1 Okta user credentials that are already assigned to this application.

If you are able to log in and see the landing page, then all your configurations up to this step are set correctly. You will not see any applications on this page yet.
Set up EMR Studio
In this step, we demonstrate the actions needed from the data lake administrator to set up EMR Studio enabled for trusted identity propagation and with IAM Identity Center integration. This allows users to directly access EMR Studio with their enterprise credentials.
Note: All Amazon S3 buckets (created after January 5, 2023) have encryption configured by default (Amazon S3 managed keys (SSE-S3)), and all new objects that are uploaded to an S3 bucket are automatically encrypted at rest. To use a different type of encryption, to meet your security needs, please update the default encryption configuration for the bucket. See Protecting data for server-side encryption for further details.
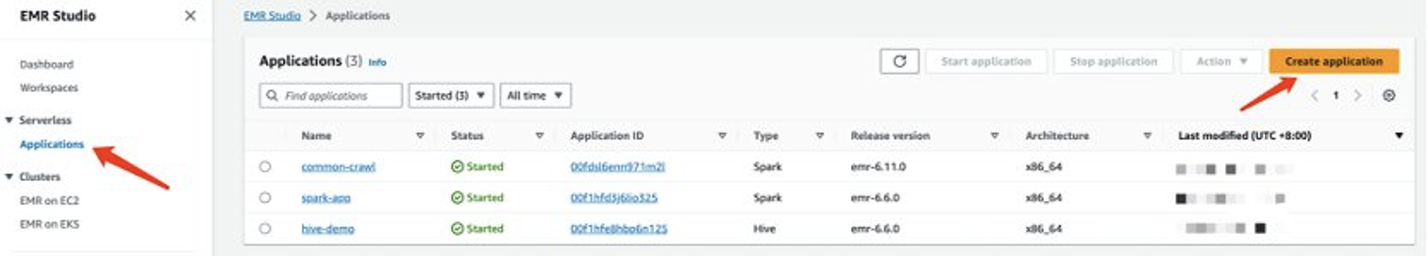
- On the Amazon EMR console, choose Studios in the navigation pane under EMR Studio.
- Choose Create Studio.

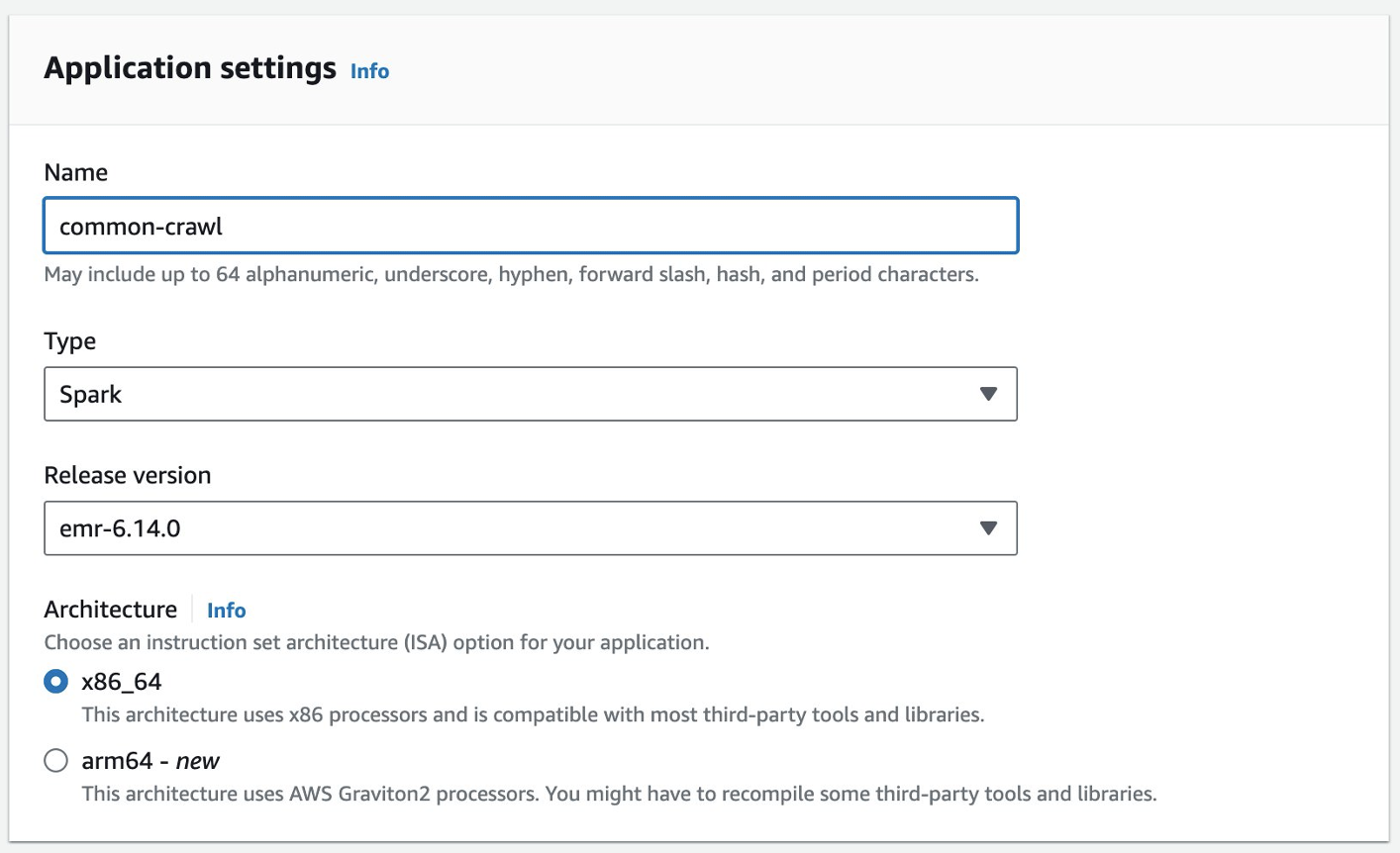
- For Setup options¸ select Custom.
- For Studio name, enter a name (for this post,
emr-studio-with-tip). - For S3 location for Workspace storage, select Select existing location and enter an existing S3 bucket (if you have one). Otherwise, select Create new bucket.

- For Service role to let Studio access your AWS resources, choose View permissions details to get the trust and IAM policy information that is needed and create a role with those specific policies in IAM. In this case, we create a new role called
emr_tip_role.

- For Service role to let Studio access your AWS resources, choose the IAM role you created.
- For Workspace name, enter a name (for this post,
studio-workspace-with-tip).

- For Authentication, select IAM Identity Center.
- For User role¸ you can create a new role or choose an existing role. For this post, we choose the role we created (
emr_tip_role). - To use the same role, add the following statement to the trust policy of the service role:
- Select Enable trusted identity propagation to allow you to control and log user access across connected applications.

- For Choose who can access your application, select All users and groups.
Later, we restrict access to resources using Lake Formation. However, there is an option here to restrict access to only assigned users and groups.
- In the Networking and security section, you can provide optional details for your VPC, subnets, and security group settings.
- Choose Create Studio.

- On the Studios page of the Amazon EMR console, locate your Studio enabled with IAM Identity Center.
- Copy the link for Studio Access URL.

- Enter the URL into a web browser and log in using Okta credentials.
You should be able to successfully sign in to the EMR Studio console.

Create an AWS Identity Center enabled security configuration for EMR clusters
EMR security configurations allow you to configure data encryption, Kerberos authentication, and Amazon S3 authorization for the EMR File System (EMRFS) on the clusters. The security configuration is available to use and reuse when you create clusters.
To integrate Amazon EMR with IAM Identity Center, you need to first create an IAM role that authenticates with IAM Identity Center from the EMR cluster. Amazon EMR uses IAM credentials to relay the IAM Identity Center identity to downstream services such as Lake Formation. The IAM role should also have the respective permissions to invoke the downstream services.
- Create a role (for this post, called
emr-idc-application) with the following trust and permission policy. The role referenced in the trust policy is theInstanceProfilerole for EMR clusters. This allows the EC2 instance profile to assume this role and act as an identity broker on behalf of the federated users.
Next, you create certificates for encrypting data in transit with Amazon EMR.
- For this post, we use OpenSSL to generate a self-signed X.509 certificate with a 2048-bit RSA private key.
The key allows access to the issuer’s EMR cluster instances in the AWS Region being used. For a complete guide on creating and providing a certificate, refer to Providing certificates for encrypting data in transit with Amazon EMR encryption.
- Upload
my-certs.zipto an S3 location that will be used to create the security configuration.
The EMR service role should have access to the S3 location. The key allows access to the issuer’s EMR cluster instances in the us-west-2 Region as specified by the *.us-west-2.compute.internal domain name as the common name. You can change this to the Region your cluster is in.

- Create an EMR security configuration with IAM Identity Center enabled from the AWS Command Line Interface (AWS CLI) with the following code:
You can view the security configuration on the Amazon EMR console.
Create a Service Catalog product template to create EMR clusters
EMR Studio with trusted identity propagation enabled can only work with clusters created from a template. Complete the following steps to create a product template in Service Catalog:
- On the Service Catalog console, choose Portfolios under Administration in the navigation pane.
- Choose Create portfolio.

- Enter a name for your portfolio (for this post,
EMR Clusters Template) and an optional description. - Choose Create.

- On the Portfolios page, choose the portfolio you just created to view its details.

- On the Products tab, choose Create product.

- For Product type, select CloudFormation.
- For Product name, enter a name (for this post,
EMR-7.0.0). - Use the security configuration
IdentityCenterConfiguration-with-lf-tipyou created in previous steps with the appropriate Amazon EMR service roles. - Choose Create product.

The following is an example CloudFormation template. Update the account-specific values for SecurityConfiguration, JobFlowRole, ServiceRole, LogUri, Ec2KeyName, and Ec2SubnetId. We provide a sample Amazon EMR service role and trust policy in Appendix A at the end of this post.
Trusted identity propagation is supported from Amazon EMR 6.15 onwards. For Amazon EMR 6.15, add the following bootstrap action to the CloudFormation script:
The portfolio now should have the EMR cluster creation product added.

- Grant the EMR Studio role
emr_tip_roleaccess to the portfolio.
Grant Lake Formation permissions to users to access data
In this step, we enable Lake Formation integration with IAM Identity Center and grant permissions to the Identity Center user analyst1. If Lake Formation is not already enabled, refer to Getting started with Lake Formation.
To use Lake Formation with Amazon EMR, create a custom role to register S3 locations. You need to create a new custom role with Amazon S3 access and not use the default role AWSServiceRoleForLakeFormationDataAccess. Additionally, enable external data filtering in Lake Formation. For more details, refer to Enable Lake Formation with Amazon EMR.
Complete the following steps to manage access permissions in Lake Formation:
- On the Lake Formation console, choose IAM Identity Center integration under Administration in the navigation pane.
Lake Formation will automatically specify the correct IAM Identity Center instance.
- Choose Create.

You can now view the IAM Identity Center integration details.

For this post, we have a Marketing database and a customer table on which we grant access to our enterprise user analyst1. You can use an existing database and table in your account or create a new one. For more examples, refer to Tutorials.
The following screenshot shows the details of our customer table.

Complete the following steps to grant analyst1 permissions. For more information, refer to Granting table permissions using the named resource method.
- On the Lake Formation console, choose Data lake permissions under Permissions in the navigation pane.
- Choose Grant.

- Select Named Data Catalog resources.
- For Databases, choose your database (
marketing). - For Tables, choose your table (
customer).

- For Table permissions, select Select and Describe.
- For Data permissions, select All data access.
- Choose Grant.

The following screenshot shows a summary of permissions that user analyst1 has. They have Select access on the table and Describe permissions on the databases.
Test the solution
To test the solution, we log in to EMR Studio as enterprise user analyst1, create a new Workspace, create an EMR cluster using a template, and use that cluster to perform an analysis. You could also use the Workspace that was created during the Studio setup. In this demonstration, we create a new Workspace.
You need additional permissions in the EMR Studio role to create and list Workspaces, use a template, and create EMR clusters. For more details, refer to Configure EMR Studio user permissions for Amazon EC2 or Amazon EKS. Appendix B at the end of this post contains a sample policy.

When the cluster is available, we attach the cluster to the Workspace and run queries on the customer table, which the user has access to.

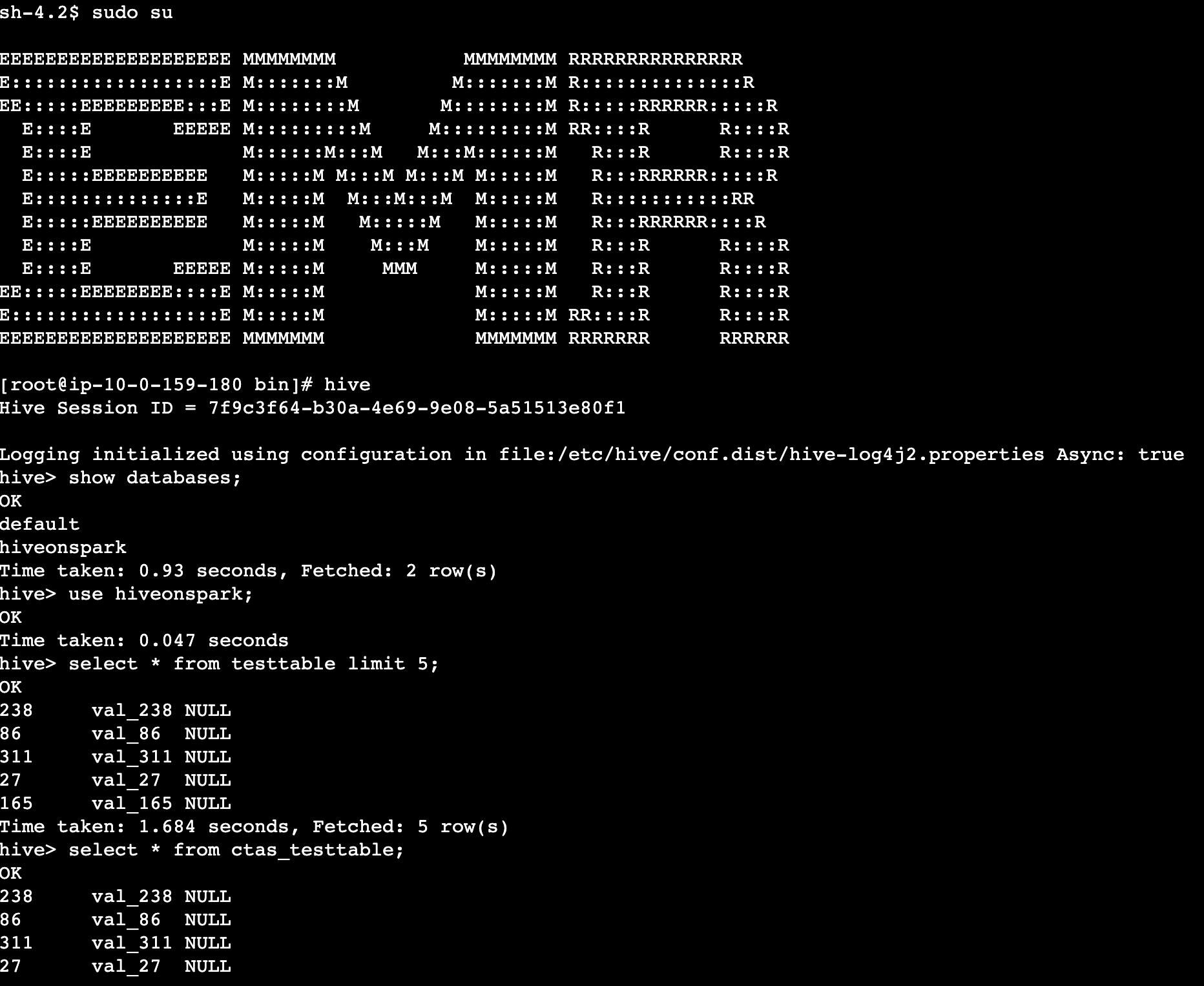
User analyst1 is now able to run queries for business use cases using their corporate identity. To open a PySpark notebook, we choose PySpark under Notebook.

When the notebook is open, we run a Spark SQL query to list the databases:

In this case, we query the customer table in the marketing database. We should be able to access the data.
Audit data access
Lake Formation API actions are logged by CloudTrail. The GetDataAccess action is logged whenever a principal or integrated AWS service requests temporary credentials to access data in a data lake location that is registered with Lake Formation. With trusted identity propagation, CloudTrail also logs the IAM Identity Center user ID of the corporate identity who requested access to the data.
The following screenshot shows the details for the analyst1 user.

Choose View event to view the event logs.

The following is an example of the GetDataAccess event log. We can trace that user analyst1, Identity Center user ID c8c11390-00a1-706e-0c7a-bbcc5a1c9a7f, has accessed the customer table.
Here is an end to end demonstration video of steps to follow for enabling trusted identity propagation to your analytics flow in Amazon EMR
Clean up
Clean up the following resources when you’re done using this solution:
- Delete the CloudFormation stacks created in each account to delete the EMR cluster.
- Delete the EMR Studio Workspaces and environment.
- Delete the Service Catalog product and portfolio.
- Delete Okta users
- Revoke Lake Formation access to the users.
Conclusion
In this post, we demonstrated how to set up and use trusted identity propagation using IAM Identity Center, EMR Studio, and Lake Formation for analytics. With trusted identity propagation, a user’s corporate identity is seamlessly propagated as they access data using single sign-on across AWS analytics services to build analytics applications. Data administrators can provide fine-grained data access directly to corporate users and groups and audit usage. To learn more, see Integrate Amazon EMR with AWS IAM Identity Center.
About the Authors
 Pradeep Misra is a Principal Analytics Solutions Architect at AWS. He works across Amazon to architect and design modern distributed analytics and AI/ML platform solutions. He is passionate about solving customer challenges using data, analytics, and AI/ML. Outside of work, Pradeep likes exploring new places, trying new cuisines, and playing board games with his family. He also likes doing science experiments with his daughters.
Pradeep Misra is a Principal Analytics Solutions Architect at AWS. He works across Amazon to architect and design modern distributed analytics and AI/ML platform solutions. He is passionate about solving customer challenges using data, analytics, and AI/ML. Outside of work, Pradeep likes exploring new places, trying new cuisines, and playing board games with his family. He also likes doing science experiments with his daughters.
 Deepmala Agarwal works as an AWS Data Specialist Solutions Architect. She is passionate about helping customers build out scalable, distributed, and data-driven solutions on AWS. When not at work, Deepmala likes spending time with family, walking, listening to music, watching movies, and cooking!
Deepmala Agarwal works as an AWS Data Specialist Solutions Architect. She is passionate about helping customers build out scalable, distributed, and data-driven solutions on AWS. When not at work, Deepmala likes spending time with family, walking, listening to music, watching movies, and cooking!
 Abhilash Nagilla is a Senior Specialist Solutions Architect at Amazon Web Services (AWS), helping public sector customers on their cloud journey with a focus on AWS analytics services. Outside of work, Abhilash enjoys learning new technologies, watching movies, and visiting new places.
Abhilash Nagilla is a Senior Specialist Solutions Architect at Amazon Web Services (AWS), helping public sector customers on their cloud journey with a focus on AWS analytics services. Outside of work, Abhilash enjoys learning new technologies, watching movies, and visiting new places.
Appendix A
Sample Amazon EMR service role and trust policy:
Note: This is a sample service role. Fine grained access control is done using Lake Formation. Modify the permissions as per your enterprise guidance and to comply with your security team.
Trust policy:
Permission Policy:
Appendix B
Sample EMR Studio role policy:
Note: This is a sample service role. Fine grained access control is done using Lake Formation. Modify the permissions as per your enterprise guidance and to comply with your security team.















![Job[14]: showString at NativeMethodAccessorImpl.java:0 and Job[15]: showString at NativeMethodAccessorImpl.java:0](https://d2908q01vomqb2.cloudfront.net/b6692ea5df920cad691c20319a6fffd7a4a766b8/2024/04/05/BDB-3979-image025.png)





 Sekar Srinivasan is a Principal Specialist Solutions Architect at AWS focused on Data Analytics and AI. Sekar has over 20 years of experience working with data. He is passionate about helping customers build scalable solutions modernizing their architecture and generating insights from their data. In his spare time he likes to work on non-profit projects, focused on underprivileged Children’s education.
Sekar Srinivasan is a Principal Specialist Solutions Architect at AWS focused on Data Analytics and AI. Sekar has over 20 years of experience working with data. He is passionate about helping customers build scalable solutions modernizing their architecture and generating insights from their data. In his spare time he likes to work on non-profit projects, focused on underprivileged Children’s education. Disha Umarwani is a Sr. Data Architect with Amazon Professional Services within Global Health Care and LifeSciences. She has worked with customers to design, architect and implement Data Strategy at scale. She specializes in architecting Data Mesh architectures for Enterprise platforms.
Disha Umarwani is a Sr. Data Architect with Amazon Professional Services within Global Health Care and LifeSciences. She has worked with customers to design, architect and implement Data Strategy at scale. She specializes in architecting Data Mesh architectures for Enterprise platforms.

















 Jeeshan Khetrapal is a Sr. Software Development Engineer at Amazon, where he develops fintech products based on cloud computing serverless architectures that are responsible for companies’ IT general controls, financial reporting, and controllership for governance, risk, and compliance.
Jeeshan Khetrapal is a Sr. Software Development Engineer at Amazon, where he develops fintech products based on cloud computing serverless architectures that are responsible for companies’ IT general controls, financial reporting, and controllership for governance, risk, and compliance. Sakti Mishra is a Principal Solutions Architect at AWS, where he helps customers modernize their data architecture and define their end-to-end data strategy, including data security, accessibility, governance, and more. He is also the author of the book
Sakti Mishra is a Principal Solutions Architect at AWS, where he helps customers modernize their data architecture and define their end-to-end data strategy, including data security, accessibility, governance, and more. He is also the author of the book 














 Brandon Abear is a Principal Data Engineer in the Data & Analytics (DnA) organization at GoDaddy. He enjoys all things big data. In his spare time, he enjoys traveling, watching movies, and playing rhythm games.
Brandon Abear is a Principal Data Engineer in the Data & Analytics (DnA) organization at GoDaddy. He enjoys all things big data. In his spare time, he enjoys traveling, watching movies, and playing rhythm games. Dinesh Sharma is a Principal Data Engineer in the Data & Analytics (DnA) organization at GoDaddy. He is passionate about user experience and developer productivity, always looking for ways to optimize engineering processes and saving cost. In his spare time, he loves reading and is an avid manga fan.
Dinesh Sharma is a Principal Data Engineer in the Data & Analytics (DnA) organization at GoDaddy. He is passionate about user experience and developer productivity, always looking for ways to optimize engineering processes and saving cost. In his spare time, he loves reading and is an avid manga fan. John Bush is a Principal Software Engineer in the Data & Analytics (DnA) organization at GoDaddy. He is passionate about making it easier for organizations to manage data and use it to drive their businesses forward. In his spare time, he loves hiking, camping, and riding his ebike.
John Bush is a Principal Software Engineer in the Data & Analytics (DnA) organization at GoDaddy. He is passionate about making it easier for organizations to manage data and use it to drive their businesses forward. In his spare time, he loves hiking, camping, and riding his ebike. Ozcan Ilikhan is the Director of Engineering for the Data and ML Platform at GoDaddy. He has over two decades of multidisciplinary leadership experience, spanning startups to global enterprises. He has a passion for leveraging data and AI in creating solutions that delight customers, empower them to achieve more, and boost operational efficiency. Outside of his professional life, he enjoys reading, hiking, gardening, volunteering, and embarking on DIY projects.
Ozcan Ilikhan is the Director of Engineering for the Data and ML Platform at GoDaddy. He has over two decades of multidisciplinary leadership experience, spanning startups to global enterprises. He has a passion for leveraging data and AI in creating solutions that delight customers, empower them to achieve more, and boost operational efficiency. Outside of his professional life, he enjoys reading, hiking, gardening, volunteering, and embarking on DIY projects. Harsh Vardhan is an AWS Solutions Architect, specializing in big data and analytics. He has over 8 years of experience working in the field of big data and data science. He is passionate about helping customers adopt best practices and discover insights from their data.
Harsh Vardhan is an AWS Solutions Architect, specializing in big data and analytics. He has over 8 years of experience working in the field of big data and data science. He is passionate about helping customers adopt best practices and discover insights from their data.



















 Edvin Hallvaxhiu is a Senior Global Security Architect with AWS Professional Services and is passionate about cybersecurity and automation. He helps customers build secure and compliant solutions in the cloud. Outside work, he likes traveling and sports.
Edvin Hallvaxhiu is a Senior Global Security Architect with AWS Professional Services and is passionate about cybersecurity and automation. He helps customers build secure and compliant solutions in the cloud. Outside work, he likes traveling and sports. Rahul Shaurya is a Principal Big Data Architect with AWS Professional Services. He helps and works closely with customers building data platforms and analytical applications on AWS. Outside of work, Rahul loves taking long walks with his dog Barney.
Rahul Shaurya is a Principal Big Data Architect with AWS Professional Services. He helps and works closely with customers building data platforms and analytical applications on AWS. Outside of work, Rahul loves taking long walks with his dog Barney. Andrea Montanari is a Senior Big Data Architect with AWS Professional Services. He actively supports customers and partners in building analytics solutions at scale on AWS.
Andrea Montanari is a Senior Big Data Architect with AWS Professional Services. He actively supports customers and partners in building analytics solutions at scale on AWS. María Guerra is a Big Data Architect with AWS Professional Services. Maria has a background in data analytics and mechanical engineering. She helps customers architecting and developing data related workloads in the cloud.
María Guerra is a Big Data Architect with AWS Professional Services. Maria has a background in data analytics and mechanical engineering. She helps customers architecting and developing data related workloads in the cloud. Pushpraj Singh is a Senior Data Architect with AWS Professional Services. He is passionate about Data and DevOps engineering. He helps customers build data driven applications at scale.
Pushpraj Singh is a Senior Data Architect with AWS Professional Services. He is passionate about Data and DevOps engineering. He helps customers build data driven applications at scale.


















 Manjit Chakraborty is a Senior Solutions Architect at AWS. He is a Seasoned & Result driven professional with extensive experience in Financial domain having worked with customers on advising, designing, leading, and implementing core-business enterprise solutions across the globe. In his spare time, Manjit enjoys fishing, practicing martial arts and playing with his daughter.
Manjit Chakraborty is a Senior Solutions Architect at AWS. He is a Seasoned & Result driven professional with extensive experience in Financial domain having worked with customers on advising, designing, leading, and implementing core-business enterprise solutions across the globe. In his spare time, Manjit enjoys fishing, practicing martial arts and playing with his daughter. Neeraj Roy is a Principal Solutions Architect at AWS based out of London. He works with Global Financial Services customers to accelerate their AWS journey. In his spare time, he enjoys reading and spending time with his family.
Neeraj Roy is a Principal Solutions Architect at AWS based out of London. He works with Global Financial Services customers to accelerate their AWS journey. In his spare time, he enjoys reading and spending time with his family.




















 Stefano Sandona is a Senior Big Data Solution Architect at AWS. He loves data, distributed systems and security. He helps customers around the world architecting secure, scalable and reliable big data platforms.
Stefano Sandona is a Senior Big Data Solution Architect at AWS. He loves data, distributed systems and security. He helps customers around the world architecting secure, scalable and reliable big data platforms. Adnan Hemani is a Software Development Engineer at AWS working with the EMR team. He focuses on the security posture of applications running on EMR clusters. He is interested in modern Big Data applications and how customers interact with them.
Adnan Hemani is a Software Development Engineer at AWS working with the EMR team. He focuses on the security posture of applications running on EMR clusters. He is interested in modern Big Data applications and how customers interact with them.

 Shijian Tang is a Analytics Specialist Solution Architect at Amazon Web Services.
Shijian Tang is a Analytics Specialist Solution Architect at Amazon Web Services. Matthew Liem is a Senior Solution Architecture Manager at Amazon Web Services.
Matthew Liem is a Senior Solution Architecture Manager at Amazon Web Services. Dalei Xu is a Analytics Specialist Solution Architect at Amazon Web Services.
Dalei Xu is a Analytics Specialist Solution Architect at Amazon Web Services. Yuanjun Xiao is a Senior Solution Architect at Amazon Web Services.
Yuanjun Xiao is a Senior Solution Architect at Amazon Web Services.



























 Raymond Lai is a Senior Solutions Architect who specializes in catering to the needs of large enterprise customers. His expertise lies in assisting customers with migrating intricate enterprise systems and databases to AWS, constructing enterprise data warehousing and data lake platforms. Raymond excels in identifying and designing solutions for AI/ML use cases, and he has a particular focus on AWS Serverless solutions and Event Driven Architecture design.
Raymond Lai is a Senior Solutions Architect who specializes in catering to the needs of large enterprise customers. His expertise lies in assisting customers with migrating intricate enterprise systems and databases to AWS, constructing enterprise data warehousing and data lake platforms. Raymond excels in identifying and designing solutions for AI/ML use cases, and he has a particular focus on AWS Serverless solutions and Event Driven Architecture design. Bin Wang, PhD, is a Senior Analytic Specialist Solutions Architect at AWS, boasting over 12 years of experience in the ML industry, with a particular focus on advertising. He possesses expertise in natural language processing (NLP), recommender systems, diverse ML algorithms, and ML operations. He is deeply passionate about applying ML/DL and big data techniques to solve real-world problems.
Bin Wang, PhD, is a Senior Analytic Specialist Solutions Architect at AWS, boasting over 12 years of experience in the ML industry, with a particular focus on advertising. He possesses expertise in natural language processing (NLP), recommender systems, diverse ML algorithms, and ML operations. He is deeply passionate about applying ML/DL and big data techniques to solve real-world problems. Aditya Shah is a Software Development Engineer at AWS. He is interested in Databases and Data warehouse engines and has worked on performance optimisations, security compliance and ACID compliance for engines like Apache Hive and Apache Spark.
Aditya Shah is a Software Development Engineer at AWS. He is interested in Databases and Data warehouse engines and has worked on performance optimisations, security compliance and ACID compliance for engines like Apache Hive and Apache Spark. Melody Yang is a Senior Big Data Solution Architect for Amazon EMR at AWS. She is an experienced analytics leader working with AWS customers to provide best practice guidance and technical advice in order to assist their success in data transformation. Her areas of interests are open-source frameworks and automation, data engineering and DataOps.
Melody Yang is a Senior Big Data Solution Architect for Amazon EMR at AWS. She is an experienced analytics leader working with AWS customers to provide best practice guidance and technical advice in order to assist their success in data transformation. Her areas of interests are open-source frameworks and automation, data engineering and DataOps.






















 Rahul Sonawane is a Principal Analytics Solutions Architect at AWS with AI/ML and Analytics as his area of specialty.
Rahul Sonawane is a Principal Analytics Solutions Architect at AWS with AI/ML and Analytics as his area of specialty. Gaurav Parekh is a Solutions Architect helping AWS customers build large scale modern architecture. He specializes in data analytics and networking. Outside of work, Gaurav enjoys playing cricket, soccer and volleyball.
Gaurav Parekh is a Solutions Architect helping AWS customers build large scale modern architecture. He specializes in data analytics and networking. Outside of work, Gaurav enjoys playing cricket, soccer and volleyball.




 Damon Cortesi is a Principal Developer Advocate with Amazon Web Services. He builds tools and content to help make the lives of data engineers easier. When not hard at work, he still builds data pipelines and splits logs in his spare time.
Damon Cortesi is a Principal Developer Advocate with Amazon Web Services. He builds tools and content to help make the lives of data engineers easier. When not hard at work, he still builds data pipelines and splits logs in his spare time.






 Saurabh Bhutyani is a Principal Analytics Specialist Solutions Architect at AWS. He is passionate about new technologies. He joined AWS in 2019 and works with customers to provide architectural guidance for running generative AI use cases, scalable analytics solutions and data mesh architectures using AWS services like Amazon Bedrock, Amazon SageMaker, Amazon EMR, Amazon Athena, AWS Glue, AWS Lake Formation, and Amazon DataZone.
Saurabh Bhutyani is a Principal Analytics Specialist Solutions Architect at AWS. He is passionate about new technologies. He joined AWS in 2019 and works with customers to provide architectural guidance for running generative AI use cases, scalable analytics solutions and data mesh architectures using AWS services like Amazon Bedrock, Amazon SageMaker, Amazon EMR, Amazon Athena, AWS Glue, AWS Lake Formation, and Amazon DataZone. Harsh Vardhan is an AWS Senior Solutions Architect, specializing in analytics. He has over 8 years of experience working in the field of big data and data science. He is passionate about helping customers adopt best practices and discover insights from their data.
Harsh Vardhan is an AWS Senior Solutions Architect, specializing in analytics. He has over 8 years of experience working in the field of big data and data science. He is passionate about helping customers adopt best practices and discover insights from their data.























 Rahul Sarda is a Senior Analytics & ML Specialist at AWS. He is a seasoned leader with over 20 years of experience, who is passionate about helping customers build scalable data and analytics solutions to gain timely insights and make critical business decisions. In his spare time, he enjoys spending time with his family, stay healthy, running and road cycling.
Rahul Sarda is a Senior Analytics & ML Specialist at AWS. He is a seasoned leader with over 20 years of experience, who is passionate about helping customers build scalable data and analytics solutions to gain timely insights and make critical business decisions. In his spare time, he enjoys spending time with his family, stay healthy, running and road cycling. Varun Rao Bhamidimarri is a Sr Manager, AWS Analytics Specialist Solutions Architect team. His focus is helping customers with adoption of cloud-enabled analytics solutions to meet their business requirements. Outside of work, he loves spending time with his wife and two kids, stay healthy, mediate and recently picked up gardening during the lockdown.
Varun Rao Bhamidimarri is a Sr Manager, AWS Analytics Specialist Solutions Architect team. His focus is helping customers with adoption of cloud-enabled analytics solutions to meet their business requirements. Outside of work, he loves spending time with his wife and two kids, stay healthy, mediate and recently picked up gardening during the lockdown.














 Ashley Zhou is a Software Development Engineer at AWS. She is interested in data analytics and distributed systems.
Ashley Zhou is a Software Development Engineer at AWS. She is interested in data analytics and distributed systems. Srividya Parthasarathy is a Senior Big Data Architect on the AWS Lake Formation team. She enjoys building analytics and data mesh solutions on AWS and sharing them with the community.
Srividya Parthasarathy is a Senior Big Data Architect on the AWS Lake Formation team. She enjoys building analytics and data mesh solutions on AWS and sharing them with the community.
 Mukul Sharma is a Software Development Engineer on Data & Analytics (DnA) organization at GoDaddy. He is a polyglot programmer with experience in a wide array of technologies to rapidly deliver scalable solutions. He enjoys singing karaoke, playing various board games, and working on personal programming projects in his spare time.
Mukul Sharma is a Software Development Engineer on Data & Analytics (DnA) organization at GoDaddy. He is a polyglot programmer with experience in a wide array of technologies to rapidly deliver scalable solutions. He enjoys singing karaoke, playing various board games, and working on personal programming projects in his spare time. Ozcan Ilikhan is a Director of Engineering on Data & Analytics (DnA) organization at GoDaddy. He is passionate about solving customer problems and increasing efficiency using data and ML/AI. In his spare time, he loves reading, hiking, gardening, and working on DIY projects.
Ozcan Ilikhan is a Director of Engineering on Data & Analytics (DnA) organization at GoDaddy. He is passionate about solving customer problems and increasing efficiency using data and ML/AI. In his spare time, he loves reading, hiking, gardening, and working on DIY projects. Ramesh Kumar Venkatraman is a Senior Solutions Architect at AWS who is passionate about containers and databases. He works with AWS customers to design, deploy, and manage their AWS workloads and architectures. In his spare time, he loves to play with his two kids and follows cricket.
Ramesh Kumar Venkatraman is a Senior Solutions Architect at AWS who is passionate about containers and databases. He works with AWS customers to design, deploy, and manage their AWS workloads and architectures. In his spare time, he loves to play with his two kids and follows cricket.







 Amit Maindola is a Senior Data Architect focused on big data and analytics at Amazon Web Services. He helps customers in their digital transformation journey and enables them to build highly scalable, robust, and secure cloud-based analytical solutions on AWS to gain timely insights and make critical business decisions.
Amit Maindola is a Senior Data Architect focused on big data and analytics at Amazon Web Services. He helps customers in their digital transformation journey and enables them to build highly scalable, robust, and secure cloud-based analytical solutions on AWS to gain timely insights and make critical business decisions.