Post Syndicated from Avijit Goswami original https://aws.amazon.com/blogs/big-data/apache-iceberg-optimization-solving-the-small-files-problem-in-amazon-emr/
In our previous post Improve operational efficiencies of Apache Iceberg tables built on Amazon S3 data lakes, we discussed how you can implement solutions to improve operational efficiencies of your Amazon Simple Storage Service (Amazon S3) data lake that is using the Apache Iceberg open table format and running on the Amazon EMR big data platform. Iceberg tables store metadata in manifest files. As the number of data files increase, the amount of metadata stored in these manifest files also increases, leading to longer query planning time. The query runtime also increases because it’s proportional to the number of data or metadata file read operations. Compaction is the process of combining these small data and metadata files to improve performance and reduce cost. Compaction also gets rid of deleting files by applying deletes and rewriting a new file without deleting records. Currently, Iceberg provides a compaction utility that compacts small files at a table or partition level. But this approach requires you to implement the compaction job using your preferred job scheduler or manually triggering the compaction job.
In this post, we discuss the new Iceberg feature that you can use to automatically compact small files while writing data into Iceberg tables using Spark on Amazon EMR or Amazon Athena.
Use cases for processing small files
Streaming applications are prone to creating a large number of small files, which can negatively impact the performance of subsequent processing times. For example, consider a critical Internet of Things (IoT) sensor from a cold storage facility that is continuously sending temperature and health data into an S3 data lake for downstream data processing and triggering actions like emergency maintenance. Systems of this nature generate a huge number of small objects and need attention to compact them to a more optimal size for faster reading, such as 128 MB, 256 MB, or 512 MB. In this post, we show you a streaming sensor data use case with a large number of small files and the mitigation steps using the Iceberg open table format. For more information on streaming applications on AWS, refer to Real-time Data Streaming and Analytics.

Solution overview
To compact the small files for improved performance, in this example, Amazon EMR triggers a compaction job after the write commit as a post-commit hook when defined thresholds (for example, number of commits) are met. By default, Amazon EMR waits for 10 commits to trigger the post-commit hook compaction utility.

This Iceberg event-based table management feature lets you monitor table activities during writes to make better decisions about how to manage each table differently based on events. As of this writing, only the optimize-data optimization is supported. To learn more about the available optimize data executors and catalog properties, refer to the README file in the GitHub repo.
To use the feature, you can use the iceberg-aws-event-based-table-management source code and provide the built JAR in the engine’s class-path. The following bootstrap action can place the JAR in the engine’s class-path:
Note that the Iceberg AWS event-based table management feature works with Iceberg v1.2.0 and above (available from Amazon EMR 6.11.0).
In some use cases, you may want to run the event-based compaction jobs in a different EMR cluster in order to avoid any impact to the ETL jobs running in their current EMR cluster. You can get the metadata, including the cluster ID of your current ETL workflows, from the /mnt/var/lib/info/job-flow.json file and then use a different cluster to process the event-based compactions.
The notebook examples shown in the following sections are also available in the aws-samples GitHub repo.
Prerequisite
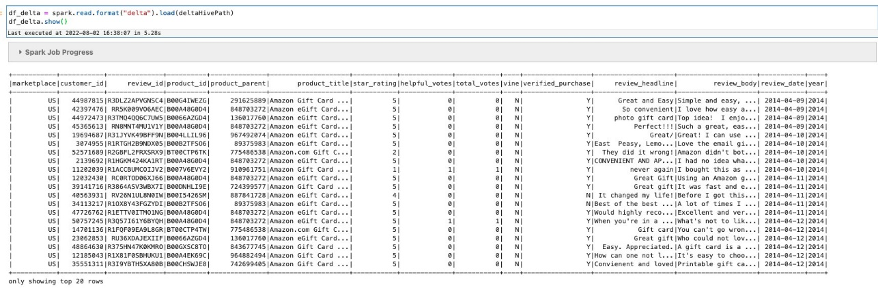
For this performance comparison exercise between a Spark external table and an Iceberg table and Iceberg with compaction, we generate a significant number of small files in Parquet format and store them in an S3 bucket. We used the Amazon Kinesis Data Generator (KDG) tool to generate sample sensor data information using the following template:
We configured an Amazon Kinesis Data Firehose delivery stream and sent the generated data into a staging S3 bucket. Then we ran an AWS Glue extract, transform, and load (ETL) job to convert the JSON files into Parquet format. For our testing, we generated about 58,176 small objects with total size of 2 GB.
For running the Amazon EMR tests, we used Amazon EMR version emr-6.11.0 with Spark 3.3.2, and JupyterEnterpriseGateway 2.6.0. The cluster used had one primary node (r5.2xlarge) and two core nodes (r5.xlarge). We used a bootstrap action during cluster creation to enable event-based table management:
Also, refer to our guidance on how to use an Iceberg cluster with Spark, which is a prerequisite for this exercise.
As part of the exercise, we see new steps are being added to the EMR cluster to trigger the compaction jobs. To enable adding new steps to the running cluster, we add the elasticmapreduce:AddJobFlowSteps action to the cluster’s default role, EMR_EC2_DefaultRole, as a prerequisite.
Performance of Iceberg reads with the compaction utility on Amazon EMR
In the following steps, we demonstrate how to use the compaction utility and what performance benefits you can achieve. We use an EMR notebook to demonstrate the benefits of the compaction utility. For instructions to set up an EMR notebook, refer to Amazon EMR Studio overview.
First, you configure your Spark session using the %%configure magic command. We use the Hive catalog for Iceberg tables.
- Before you run the following step, create an Amazon S3 bucket in your AWS account called <your-iceberg-storage-blog>. To check how to create an Amazon S3 bucket, follow the instructions given here. Update the
your-iceberg-storage-blogbucket name in the following configuration with the actual bucket name you created to test this example: - Create a new database for the Iceberg table in the AWS Glue Data Catalog named DB and provide the S3 URI specified in the Spark config as
s3://<your-iceberg-storage-blog>/iceberg/db. Also, create another Database named iceberg_db in Glue for the parquet tables. Follow the instructions given in Working with databases on the AWS Glue console to create your Glue databases. Then create a new Spark table in Parquet format pointing to the bucket containing small objects in your AWS account. See the following code: - Run an aggregate SQL to measure the performance of Spark SQL on the Parquet table with 58,176 small objects:

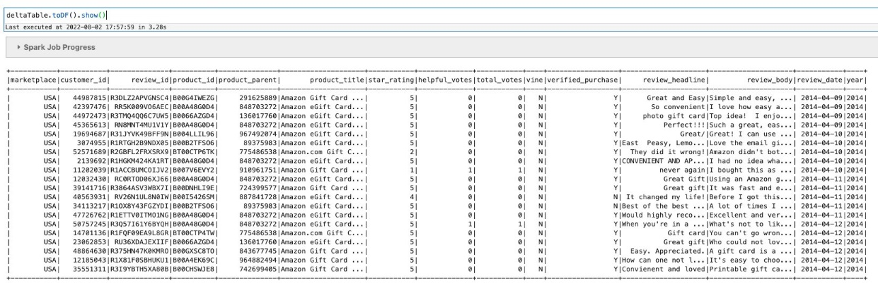
In the following steps, we create a new Iceberg table from the Spark/Parquet table using CTAS (Create Table As Select). Then we show how the automated compaction job can help improve query performance.
- Create a new Iceberg table using CTAS from the earlier AWS Glue table with the small files:
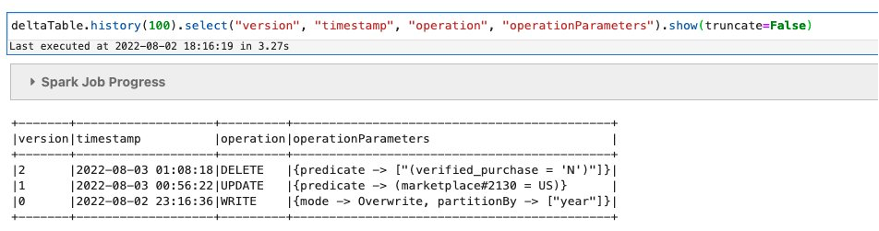
- Validate that a new Iceberg snapshot was created for the new table:

We have confirmed that our S3 folder corresponds to the newly created Iceberg table. It shows that during the CTAS statement, it added 1,879 objects in the new folder with a total size of 1.3 GB. We can conclude that Iceberg did some optimization while loading data from the Parquet table.
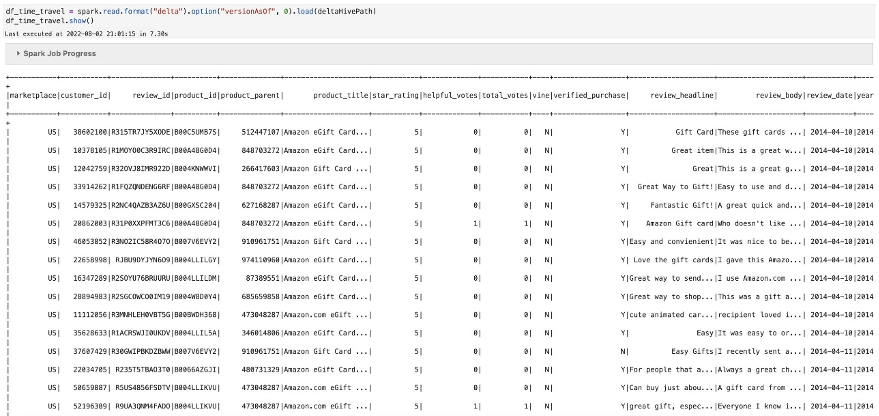
- Now that you have data in the Iceberg table, run the previous aggregation SQL to check the runtime:

The runtime for the preceding query ran on the Iceberg table with 1,879 objects in 1 minute, 39 seconds. There is already some significant performance improvement by converting the external Parquet table to an Iceberg table.
- Now let’s add the configurations needed to apply the automatic compaction of small files in the Iceberg tables. Note the last four newly added configurations in the following statement. The parameter
optimize-data.commit-thresholdsuggests that the compaction will take place after the first successful commit. The default is 10 successful commits to trigger the compaction. - Run a quick sanity check to confirm that the configurations are working fine with Spark SQL.

- 10. To activate the automatic compaction process, add a new record to the existing Iceberg table using a Spark insert:
- Navigate to the Amazon EMR console to check the cluster steps.
You should see a new step added that goes from Pending to Running and finally the Completed state. Every time the data in the Iceberg table is updated or inserted, based on configuration optimize-data.commit-threshold, the optimize job will automatically trigger to compact the underlying data.

- Validate that the record insert was successful.

- Check the snapshot table to see that a new snapshot is created for the table with the operation
replace.
For every successful run of the background optimize job, a new entry will be added to the snapshot table.

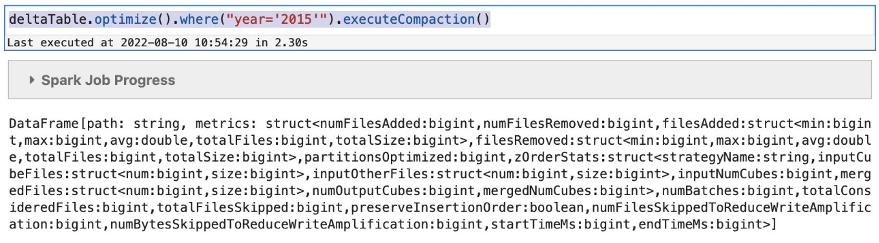
- On the Amazon S3 console, navigate to the folder corresponding to the Iceberg table and see that the data files are compacted.
In our case, it was compacted from the previous smaller sizes to approximately 437 MB. The folder will still contain the previous smaller files for time travel unless you issue an expire snapshot command to remove them.
- Now you can run the same aggregate query and record the performance after the compaction.

Summary of Amazon EMR testing
The runtime for the preceding aggregation query on the compacted Iceberg table reduced to approximately 59 seconds from the previous runtime of 1 minute, 39 seconds. That is about a 40% improvement. The more small files you have in your source bucket, the bigger performance boost you can achieve with this post-hook compaction implementation. The examples shown in this blog were executed in a small Amazon EMR cluster with only two core nodes (r5.xlarge). To improve the performance of your Spark applications, Amazon EMR provides multiple optimization features that you can implement for your production workloads.

Performance of Iceberg reads with the compaction utility on Athena
To manage the Iceberg table based on events, you can start the Spark 3.3 SQL shell as shown in the following code. Make sure that the athena:StartQueryExecution and athena:GetQueryExecution permission policies are enabled.
Clean up
After you complete the test, clean up your resources to avoid any recurring costs:
- Delete the S3 buckets that you created for this test.
- Delete the EMR cluster.
- Stop and delete the EMR notebook instance.
Conclusion
In this post, we showed how Iceberg event-based table management lets you manage each table differently based on events and compact small files to boost application performance. This event-based process significantly reduces the operational overhead of using the Iceberg rewrite_data_files procedure, which needs manual or scheduled operation.
To learn more about Apache Iceberg and implement this open table format for your transactional data lake use cases, refer to the following resources:
- Apache Iceberg table specifications
- Apache Iceberg support on Amazon EMR
- Use a cluster with Iceberg installed
- Improve operational efficiencies of Apache Iceberg tables built on Amazon S3 data lakes
About the Authors
 Avijit Goswami is a Principal Solutions Architect at AWS specialized in data and analytics. He supports AWS strategic customers in building high-performing, secure, and scalable data lake solutions on AWS using AWS managed services and open-source solutions. Outside of his work, Avijit likes to travel, hike, watch sports, and listen to music.
Avijit Goswami is a Principal Solutions Architect at AWS specialized in data and analytics. He supports AWS strategic customers in building high-performing, secure, and scalable data lake solutions on AWS using AWS managed services and open-source solutions. Outside of his work, Avijit likes to travel, hike, watch sports, and listen to music.
 Rajarshi Sarkar is a Software Development Engineer at Amazon EMR/Athena. He works on cutting-edge features of Amazon EMR/Athena and is also involved in open-source projects such as Apache Iceberg and Trino. In his spare time, he likes to travel, watch movies, and hang out with friends.
Rajarshi Sarkar is a Software Development Engineer at Amazon EMR/Athena. He works on cutting-edge features of Amazon EMR/Athena and is also involved in open-source projects such as Apache Iceberg and Trino. In his spare time, he likes to travel, watch movies, and hang out with friends.








 Rajarshi Sarkar is a Software Development Engineer at Amazon EMR/Athena. He works on cutting-edge features of Amazon EMR/Athena and is also involved in open-source projects such as Apache Iceberg and Trino. In his spare time, he likes to travel, watch movies, and hang out with friends.
Rajarshi Sarkar is a Software Development Engineer at Amazon EMR/Athena. He works on cutting-edge features of Amazon EMR/Athena and is also involved in open-source projects such as Apache Iceberg and Trino. In his spare time, he likes to travel, watch movies, and hang out with friends. Prashant Singh is a Software Development Engineer at AWS. He is interested in Databases and Data Warehouse engines and has worked on Optimizing Apache Spark performance on EMR. He is an active contributor in open source projects like Apache Spark and Apache Iceberg. During his free time, he enjoys exploring new places, food and hiking.
Prashant Singh is a Software Development Engineer at AWS. He is interested in Databases and Data Warehouse engines and has worked on Optimizing Apache Spark performance on EMR. He is an active contributor in open source projects like Apache Spark and Apache Iceberg. During his free time, he enjoys exploring new places, food and hiking.


 Avijit Goswami is a Principal Solutions Architect at AWS specialized in data and analytics. He supports AWS strategic customers in building high-performing, secure, and scalable data lake solutions on AWS using AWS managed services and open-source solutions. Outside of his work, Avijit likes to travel, hike in the San Francisco Bay Area trails, watch sports, and listen to music.
Avijit Goswami is a Principal Solutions Architect at AWS specialized in data and analytics. He supports AWS strategic customers in building high-performing, secure, and scalable data lake solutions on AWS using AWS managed services and open-source solutions. Outside of his work, Avijit likes to travel, hike in the San Francisco Bay Area trails, watch sports, and listen to music. Ajit Tandale is a Big Data Solutions Architect at Amazon Web Services. He helps AWS strategic customers accelerate their business outcomes by providing expertise in big data using AWS managed services and open-source solutions. Outside of work, he enjoys reading, biking, and watching sci-fi movies.
Ajit Tandale is a Big Data Solutions Architect at Amazon Web Services. He helps AWS strategic customers accelerate their business outcomes by providing expertise in big data using AWS managed services and open-source solutions. Outside of work, he enjoys reading, biking, and watching sci-fi movies. Thippana Vamsi Kalyan is a Software Development Engineer at AWS. He is passionate about learning and building highly scalable and reliable data analytics services and solutions on AWS. In his free time, he enjoys reading, being outdoors with his wife and kid, walking, and watching sports and movies.
Thippana Vamsi Kalyan is a Software Development Engineer at AWS. He is passionate about learning and building highly scalable and reliable data analytics services and solutions on AWS. In his free time, he enjoys reading, being outdoors with his wife and kid, walking, and watching sports and movies.




 Avijit Goswami is a Principal Solutions Architect at AWS, helping startup customers become tomorrow’s enterprises using AWS services. He is part of the Analytics Specialist community at AWS. When not at work, Avijit likes to cook, travel, hike, watch sports, and listen to music.
Avijit Goswami is a Principal Solutions Architect at AWS, helping startup customers become tomorrow’s enterprises using AWS services. He is part of the Analytics Specialist community at AWS. When not at work, Avijit likes to cook, travel, hike, watch sports, and listen to music.
 Mohit Saxena is a Technical Lead Manager at AWS Glue. His team works on Glue’s Spark runtime to enable new customer use cases for efficiently managing data lakes on AWS and optimize Apache Spark for performance and reliability.
Mohit Saxena is a Technical Lead Manager at AWS Glue. His team works on Glue’s Spark runtime to enable new customer use cases for efficiently managing data lakes on AWS and optimize Apache Spark for performance and reliability.