Post Syndicated from Vishal Pathak original https://aws.amazon.com/blogs/big-data/creating-a-source-to-lakehouse-data-replication-pipe-using-apache-hudi-aws-glue-aws-dms-and-amazon-redshift/
Most customers have their applications backed by various sql and nosql systems on prem and on cloud. Since the data is in various independent systems, customers struggle to derive meaningful info by combining data from all of these sources. Hence, customers create data lakes to bring their data in a single place.
Typically, a replication tool such as AWS Database Migration Service (AWS DMS) can replicate the data from your source systems to Amazon Simple Storage Service (Amazon S3). When the data is in Amazon S3, customers process it based on their requirements. A typical requirement is to sync the data in Amazon S3 with the updates on the source systems. Although it’s easy to apply updates on a relational database management system (RDBMS) that backs an online source application, it’s tough to apply this change data capture (CDC) process on your data lakes. Apache Hudi is a good way to solve this problem. Currently, you can use Hudi on Amazon EMR to create Hudi tables.
In this post, we use Apache Hudi to create tables in the AWS Glue Data Catalog using AWS Glue jobs. AWS Glue is a fully managed extract, transform, and load (ETL) service that makes it easy to prepare and load your data for analytics. This post enables you to take advantage of the serverless architecture of AWS Glue while upserting data in your data lake, hassle-free.
To write to Hudi tables using AWS Glue jobs, we use a JAR file created using open-source Apache Hudi. This JAR file is used as a dependency in the AWS Glue jobs created through the AWS CloudFormation template provided in this post. Steps to create the JAR file are included in the appendix.
The following diagram illustrates the architecture the CloudFormation template implements.

Prerequisites
The CloudFormation template requires you to select an Amazon Elastic Compute Cloud (Amazon EC2) key pair. This key is configured on an EC2 instance that lives in the public subnet. We use this EC2 instance to get to the Aurora cluster that lives in the private subnet. Make sure you have a key in the Region where you deploy the template. If you don’t have one, you can create a new key pair.
Solution overview
The following are the high-level implementation steps:
- Create a CloudFormation stack using the provided template.
- Connect to the Amazon Aurora cluster used as a source for this post.
- Run InitLoad_TestStep1.sql, in the source Amazon Aurora cluster, to create a schema and a table.
AWS DMS replicates the data from the Aurora cluster to the raw S3 bucket. AWS DMS supports a variety of sources.
The CloudFormation stack creates an AWS Glue job (HudiJob) that is scheduled to run at a frequency set in the ScheduleToRunGlueJob parameter of the CloudFormation stack. This job reads the data from the raw S3 bucket, writes to the Curated S3 bucket, and creates a Hudi table in the Data Catalog. The job also creates an Amazon Redshift external schema in the Amazon Redshift cluster created by the CloudFormation stack.
- You can now query the Hudi table in Amazon Athena or Amazon Redshift. Visit Creating external tables for data managed in Apache Hudi or Considerations and Limitations to query Apache Hudi datasets in Amazon Athena for details.
- Run IncrementalUpdatesAndInserts_TestStep2.sql on the source Aurora cluster.
This incremental data is also replicated to the raw S3 bucket through AWS DMS. HudiJob picks up the incremental data, using AWS Glue bookmarks, and applies it to the Hudi table created earlier.
- You can now query the changed data.
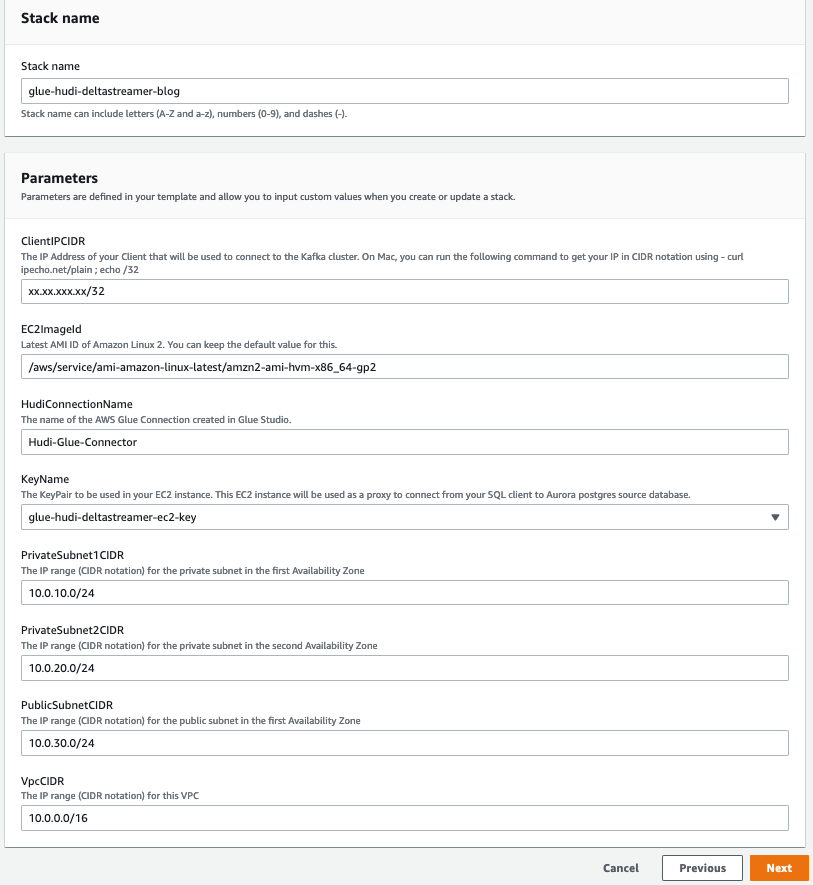
Creating your CloudFormation stack
Click on the Launch Stack button to get started and provide the following parameters:

| Parameter |
Description |
VpcCIDR |
CIDR range for the VPC. |
PrivateSubnet1CIDR |
CIDR range for the first private subnet. |
PrivateSubnet2CIDR |
CIDR range for the second private subnet. |
PublicSubnetCIDR |
CIDR range for the public subnet. |
AuroraDBMasterUserPassword |
Primary user password for the Aurora cluster. |
RedshiftDWMasterUserPassword |
Primary user password for the Amazon Redshift data warehouse. |
KeyName |
The EC2 key pair to be configured in the EC2 instance on the public subnet. This EC2 instance is used to get to the Aurora cluster in the private subnet. Select the value from the dropdown. |
ClientIPCIDR |
Your IP address in CIDR notation. The CloudFormation template creates a security group rule that grants ingress on port 22 to this IP address. On a Mac, you can run the following command to get your IP address: curl ipecho.net/plain ; echo /32 |
EC2ImageId |
The image ID used to create the EC2 instance in the public subnet to be a jump box to connect to the source Aurora cluster. If you supply your image ID, the template uses it to create the EC2 instance. |
HudiStorageType |
This is used by the AWS Glue job to determine if you want to create a CoW or MoR storage type table. Enter MoR if you want to create MoR storage type tables. |
ScheduleToRunGlueJob |
The AWS Glue job runs on a schedule to pick the new files and load to the curated bucket. This parameter sets the schedule of the job. |
DMSBatchUnloadIntervalInSecs |
AWS DMS batches the inputs from the source and loads the output to the taw bucket. This parameter defines the frequency in which the data is loaded to the raw bucket. |
GlueJobDPUs |
The number of DPUs that are assigned to the two AWS Glue jobs. |
To simplify running the template, your account is given permissions on the key used to encrypt the resources in the CloudFormation template. You can restrict that to the role if desired.
Granting Lake Formation permissions
AWS Lake Formation enables customers to set up fine grained access control for their Datalake. Detail steps to set up AWS Lake Formation can be found here.
Setting up AWS Lake Formation is out of scope for this post. However, if you have Lake Formation configured in the Region where you’re deploying this template, grant Create database permission to the LakeHouseExecuteGlueHudiJobRole role after the CloudFormation stack is successfully created.

This will ensure that you don’t get the following error while running your AWS Glue job.
org.apache.spark.sql.AnalysisException: org.apache.hadoop.hive.ql.metadata.HiveException: MetaException(message:Insufficient Lake Formation permission(s) on global_temp
Similarly grant Describe permission to the LakeHouseExecuteGlueHudiJobRole role on default database.


This will ensure that you don’t get the following error while running your AWS Glue job.
AnalysisException: 'java.lang.RuntimeException: MetaException(message:Unable to verify existence of default database: com.amazonaws.services.glue.model.AccessDeniedException: Insufficient Lake Formation permission(s) on default (Service: AWSGlue; Status Code: 400; Error Code: AccessDeniedException;
Connecting to source Aurora cluster
To connect to source Aurora cluster using SQL Workbench, complete the following steps:
- On SQL Workbench, under File, choose Connect window.

- Choose Manage Drivers.

- Choose PostgreSQL.
- For Library, use the driver JAR file.
- For Classname, enter
org.postgresql.Driver.
- For Sample URL, enter
jdbc:postgresql://host:port/name_of_database.

- Click the Create a new connection profile button.
- For Driver, choose your new PostgreSQL driver.
- For URL, enter
lakehouse_source_db after port/.
- For Username, enter
postgres.
- For Password, enter the same password that you used for the
AuroraDBMasterUserPassword parameter while creating the CloudFormation stack.
- Choose SSH.

- On the Outputs tab of your CloudFormation stack, copy the IP address next to
PublicIPOfEC2InstanceForTunnel and enter it for SSH hostname.

- For SSH port, enter
22.
- For Username, enter
ec2-user.
- For Private key file, enter the private key for the public key chosen in the
KeyName parameter of the CloudFormation stack.
- For Local port, enter any available local port number.
- On the Outputs tab of your stack, copy the value next to
EndpointOfAuroraCluster and enter it for DB hostname.
- For DB port, enter
5432.
- Select Rewrite JDBC URL.

Checking the Rewrite JDBC URL checkbox will automatically feed in the value of host and port in the URL text box as shown below.
- Test the connection and make sure that you get a message that the connection was successful.

Troubleshooting
Complete the following steps if you receive this message: Could not initialize SSH tunnel: java.net.ConnectException: Operation timed out (Connection timed out)
- Go to your CloudFormation stack and search for
LakeHouseSecurityGroup under Resources .
- Choose the link in the Physical ID.

- Select your security group.
- From the Actions menu, choose Edit inbound rules.

- Look for the rule with the description:
Rule to allow connection from the SQL client to the EC2 instance used as jump box for SSH tunnel
- From the Source menu, choose My IP.
- Choose Save rules.

- Test the connection from your SQL Workbench again and make sure that you get a successful message.
Running the initial load script
You’re now ready to run the InitLoad_TestStep1.sql script to create some test data.
- Open InitLoad_TestStep1.sql in your SQL client and run it.

The output shows that 11 statements have been run.
AWS DMS replicates these inserts to your raw S3 bucket at the frequency set in the DMSBatchUnloadIntervalInSecs parameter of your CloudFormation stack.
- On the AWS DMS console, choose the
lakehouse-aurora-src-to-raw-s3-tgt task:
- On the Table statistics tab, you should see the seven full load rows of
employee_details have been replicated.

The lakehouse-aurora-src-to-raw-s3-tgt replication task has the following table mapping with transformation to add a schema name and a table name as additional columns:
{
"rules":[
{
"rule-type":"selection",
"rule-id":"1",
"rule-name":"1",
"object-locator":{
"schema-name":"human_resources",
"table-name":"%"
},
"rule-action":"include",
"filters":[
]
},
{
"rule-type":"transformation",
"rule-id":"2",
"rule-name":"2",
"rule-target":"column",
"object-locator":{
"schema-name":"%",
"table-name":"%"
},
"rule-action":"add-column",
"value":"schema_name",
"expression":"$SCHEMA_NAME_VAR",
"data-type":{
"type":"string",
"length":50
}
},
{
"rule-type":"transformation",
"rule-id":"3",
"rule-name":"3",
"rule-target":"column",
"object-locator":{
"schema-name":"%",
"table-name":"%"
},
"rule-action":"add-column",
"value":"table_name",
"expression":"$TABLE_NAME_VAR",
"data-type":{
"type":"string",
"length":50
}
}
]
}
These settings put the name of the source schema and table as two additional columns in the output Parquet file of AWS DMS.
These columns are used in the AWS Glue HudiJob to find out the tables that have new inserts, updates, or deletes.
- On the Resources tab of the CloudFormation stack, locate
RawS3Bucket.
- Choose the Physical ID link.

- Navigate to
human_resources/employee_details.
The LOAD00000001.parquet file is created under human_resources/employee_details. (The name of your raw bucket is different from the following screenshot).

You can also see the time of creation of this file. You should have at least one successful run of the AWS Glue job (HudiJob) after this time for the Hudi table to be created. The AWS Glue job is configured to load this data into the curated bucket at the frequency set in the ScheduleToRunGlueJob parameter of your CloudFormation stack. The default is 5 minutes.
AWS Glue job HudiJob
The following code is the script for HudiJob:
import sys
import os
import json
from pyspark.context import SparkContext
from pyspark.sql.session import SparkSession
from pyspark.sql.functions import concat, col, lit, to_timestamp
from awsglue.utils import getResolvedOptions
from awsglue.context import GlueContext
from awsglue.job import Job
from awsglue.dynamicframe import DynamicFrame
import boto3
from botocore.exceptions import ClientError
args = getResolvedOptions(sys.argv, ['JOB_NAME'])
spark = SparkSession.builder.config('spark.serializer','org.apache.spark.serializer.KryoSerializer').getOrCreate()
glueContext = GlueContext(spark.sparkContext)
job = Job(glueContext)
job.init(args['JOB_NAME'], args)
logger = glueContext.get_logger()
logger.info('Initialization.')
glueClient = boto3.client('glue')
ssmClient = boto3.client('ssm')
redshiftDataClient = boto3.client('redshift-data')
logger.info('Fetching configuration.')
region = os.environ['AWS_DEFAULT_REGION']
curatedS3BucketName = ssmClient.get_parameter(Name='lakehouse-curated-s3-bucket-name')['Parameter']['Value']
rawS3BucketName = ssmClient.get_parameter(Name='lakehouse-raw-s3-bucket-name')['Parameter']['Value']
hudiStorageType = ssmClient.get_parameter(Name='lakehouse-hudi-storage-type')['Parameter']['Value']
dropColumnList = ['db','table_name','Op']
logger.info('Getting list of schema.tables that have changed.')
changeTableListDyf = glueContext.create_dynamic_frame_from_options(connection_type = 's3', connection_options = {'paths': ['s3://'+rawS3BucketName], 'groupFiles': 'inPartition', 'recurse':True}, format = 'parquet', format_options={}, transformation_ctx = 'changeTableListDyf')
logger.info('Processing starts.')
if(changeTableListDyf.count() > 0):
logger.info('Got new files to process.')
changeTableList = changeTableListDyf.toDF().select('schema_name','table_name').distinct().rdd.map(lambda row : row.asDict()).collect()
for dbName in set([d['schema_name'] for d in changeTableList]):
spark.sql('CREATE DATABASE IF NOT EXISTS ' + dbName)
redshiftDataClient.execute_statement(ClusterIdentifier='lakehouse-redshift-cluster', Database='lakehouse_dw', DbUser='rs_admin', Sql='CREATE EXTERNAL SCHEMA IF NOT EXISTS ' + dbName + ' FROM DATA CATALOG DATABASE \'' + dbName + '\' REGION \'' + region + '\' IAM_ROLE \'' + boto3.client('iam').get_role(RoleName='LakeHouseRedshiftGlueAccessRole')['Role']['Arn'] + '\'')
for i in changeTableList:
logger.info('Looping for ' + i['schema_name'] + '.' + i['table_name'])
dbName = i['schema_name']
tableNameCatalogCheck = ''
tableName = i['table_name']
if(hudiStorageType == 'MoR'):
tableNameCatalogCheck = i['table_name'] + '_ro' #Assumption is that if _ro table exists then _rt table will also exist. Hence we are checking only for _ro.
else:
tableNameCatalogCheck = i['table_name'] #The default config in the CF template is CoW. So assumption is that if the user hasn't explicitly requested to create MoR storage type table then we will create CoW tables. Again, if the user overwrites the config with any value other than 'MoR' we will create CoW storage type tables.
isTableExists = False
isPrimaryKey = False
isPartitionKey = False
primaryKey = ''
partitionKey = ''
try:
glueClient.get_table(DatabaseName=dbName,Name=tableNameCatalogCheck)
isTableExists = True
logger.info(dbName + '.' + tableNameCatalogCheck + ' exists.')
except ClientError as e:
if e.response['Error']['Code'] == 'EntityNotFoundException':
isTableExists = False
logger.info(dbName + '.' + tableNameCatalogCheck + ' does not exist. Table will be created.')
try:
table_config = json.loads(ssmClient.get_parameter(Name='lakehouse-table-' + dbName + '.' + tableName)['Parameter']['Value'])
try:
primaryKey = table_config['primaryKey']
isPrimaryKey = True
logger.info('Primary key:' + primaryKey)
except KeyError as e:
isPrimaryKey = False
logger.info('Primary key not found. An append only glueparquet table will be created.')
try:
partitionKey = table_config['partitionKey']
isPartitionKey = True
logger.info('Partition key:' + partitionKey)
except KeyError as e:
isPartitionKey = False
logger.info('Partition key not found. Partitions will not be created.')
except ClientError as e:
if e.response['Error']['Code'] == 'ParameterNotFound':
isPrimaryKey = False
isPartitionKey = False
logger.info('Config for ' + dbName + '.' + tableName + ' not found in parameter store. Non partitioned append only table will be created.')
inputDyf = glueContext.create_dynamic_frame_from_options(connection_type = 's3', connection_options = {'paths': ['s3://' + rawS3BucketName + '/' + dbName + '/' + tableName], 'groupFiles': 'none', 'recurse':True}, format = 'parquet',transformation_ctx = tableName)
inputDf = inputDyf.toDF().withColumn('update_ts_dms',to_timestamp(col('update_ts_dms')))
targetPath = 's3://' + curatedS3BucketName + '/' + dbName + '/' + tableName
morConfig = {'hoodie.datasource.write.storage.type': 'MERGE_ON_READ', 'hoodie.compact.inline': 'false', 'hoodie.compact.inline.max.delta.commits': 20, 'hoodie.parquet.small.file.limit': 0}
commonConfig = {'className' : 'org.apache.hudi', 'hoodie.datasource.hive_sync.use_jdbc':'false', 'hoodie.datasource.write.precombine.field': 'update_ts_dms', 'hoodie.datasource.write.recordkey.field': primaryKey, 'hoodie.table.name': tableName, 'hoodie.consistency.check.enabled': 'true', 'hoodie.datasource.hive_sync.database': dbName, 'hoodie.datasource.hive_sync.table': tableName, 'hoodie.datasource.hive_sync.enable': 'true'}
partitionDataConfig = {'hoodie.datasource.write.partitionpath.field': partitionKey, 'hoodie.datasource.hive_sync.partition_extractor_class': 'org.apache.hudi.hive.MultiPartKeysValueExtractor', 'hoodie.datasource.hive_sync.partition_fields': partitionKey}
unpartitionDataConfig = {'hoodie.datasource.hive_sync.partition_extractor_class': 'org.apache.hudi.hive.NonPartitionedExtractor', 'hoodie.datasource.write.keygenerator.class': 'org.apache.hudi.keygen.NonpartitionedKeyGenerator'}
incrementalConfig = {'hoodie.upsert.shuffle.parallelism': 20, 'hoodie.datasource.write.operation': 'upsert', 'hoodie.cleaner.policy': 'KEEP_LATEST_COMMITS', 'hoodie.cleaner.commits.retained': 10}
initLoadConfig = {'hoodie.bulkinsert.shuffle.parallelism': 3, 'hoodie.datasource.write.operation': 'bulk_insert'}
deleteDataConfig = {'hoodie.datasource.write.payload.class': 'org.apache.hudi.common.model.EmptyHoodieRecordPayload'}
if(hudiStorageType == 'MoR'):
commonConfig = {**commonConfig, **morConfig}
logger.info('MoR config appended to commonConfig.')
combinedConf = {}
if(isPrimaryKey):
logger.info('Going the Hudi way.')
if(isTableExists):
logger.info('Incremental load.')
outputDf = inputDf.filter("Op != 'D'").drop(*dropColumnList)
if outputDf.count() > 0:
logger.info('Upserting data.')
if (isPartitionKey):
logger.info('Writing to partitioned Hudi table.')
outputDf = outputDf.withColumn(partitionKey,concat(lit(partitionKey+'='),col(partitionKey)))
combinedConf = {**commonConfig, **partitionDataConfig, **incrementalConfig}
outputDf.write.format('org.apache.hudi').options(**combinedConf).mode('Append').save(targetPath)
else:
logger.info('Writing to unpartitioned Hudi table.')
combinedConf = {**commonConfig, **unpartitionDataConfig, **incrementalConfig}
outputDf.write.format('org.apache.hudi').options(**combinedConf).mode('Append').save(targetPath)
outputDf_deleted = inputDf.filter("Op = 'D'").drop(*dropColumnList)
if outputDf_deleted.count() > 0:
logger.info('Some data got deleted.')
if (isPartitionKey):
logger.info('Deleting from partitioned Hudi table.')
outputDf_deleted = outputDf_deleted.withColumn(partitionKey,concat(lit(partitionKey+'='),col(partitionKey)))
combinedConf = {**commonConfig, **partitionDataConfig, **incrementalConfig, **deleteDataConfig}
outputDf_deleted.write.format('org.apache.hudi').options(**combinedConf).mode('Append').save(targetPath)
else:
logger.info('Deleting from unpartitioned Hudi table.')
combinedConf = {**commonConfig, **unpartitionDataConfig, **incrementalConfig, **deleteDataConfig}
outputDf_deleted.write.format('org.apache.hudi').options(**combinedConf).mode('Append').save(targetPath)
else:
outputDf = inputDf.drop(*dropColumnList)
if outputDf.count() > 0:
logger.info('Inital load.')
if (isPartitionKey):
logger.info('Writing to partitioned Hudi table.')
outputDf = outputDf.withColumn(partitionKey,concat(lit(partitionKey+'='),col(partitionKey)))
combinedConf = {**commonConfig, **partitionDataConfig, **initLoadConfig}
outputDf.write.format('org.apache.hudi').options(**combinedConf).mode('Overwrite').save(targetPath)
else:
logger.info('Writing to unpartitioned Hudi table.')
combinedConf = {**commonConfig, **unpartitionDataConfig, **initLoadConfig}
outputDf.write.format('org.apache.hudi').options(**combinedConf).mode('Overwrite').save(targetPath)
else:
if (isPartitionKey):
logger.info('Writing to partitioned glueparquet table.')
sink = glueContext.getSink(connection_type = 's3', path= targetPath, enableUpdateCatalog = True, updateBehavior = 'UPDATE_IN_DATABASE', partitionKeys=[partitionKey])
else:
logger.info('Writing to unpartitioned glueparquet table.')
sink = glueContext.getSink(connection_type = 's3', path= targetPath, enableUpdateCatalog = True, updateBehavior = 'UPDATE_IN_DATABASE')
sink.setFormat('glueparquet')
sink.setCatalogInfo(catalogDatabase = dbName, catalogTableName = tableName)
outputDyf = DynamicFrame.fromDF(inputDf.drop(*dropColumnList), glueContext, 'outputDyf')
sink.writeFrame(outputDyf)
job.commit()
Hudi tables need a primary key to perform upserts. Hudi tables can also be partitioned based on a certain key. We get the names of the primary key and the partition key from AWS Systems Manager Parameter Store.
The HudiJob script looks for an AWS Systems Manager Parameter with the naming format lakehouse-table-<schema_name>.<table_name>. It compares the name of the parameter with the name of the schema and table columns, added by AWS DMS, to get the primary key and the partition key for the Hudi table.
The CloudFormation template creates lakehouse-table-human_resources.employee_details AWS Systems Manager Parameter, as shown on the Resources tab.

If you choose the Physical ID link, you can locate the value of the AWS Systems Manager Parameter. The AWS Systems Manager Parameter has {"primaryKey": "emp_no", "partitionKey": "department"} value in it.

Because of the value in the lakehouse-table-human_resources.employee_details AWS Systems Manager Parameter, the AWS Glue script creates a human_resources.employee_details Hudi table partitioned on the department column for the employee_details table created in the source using the InitLoad_TestStep1.sql script. The HudiJob also uses the emp_no column as the primary key for upserts.
If you reuse this CloudFormation template and create your own table, you have to create an associated AWS Systems Manager Parameter with the naming convention lakehouse-table-<schema_name>.<table_name>. Keep in mind the following:
- If you don’t create a parameter, the script creates an unpartitioned
glueparquet append-only table.
- If you create a parameter that only has the
primaryKey part in the value, the script creates an unpartitioned Hudi table.
- If you create a parameter that only has the
partitionKey part in the value, the script creates a partitioned glueparquet append-only table.
If you have too many tables to replicate, you can also store the primary key and partition key configuration in Amazon DynamoDB or Amazon S3 and change the code accordingly.
In the InitLoad_TestStep1.sql script, replica identity for human_resources.employee_details table is set to full. This makes sure that AWS DMS transfers the full delete record to Amazon S3. Having this delete record is important for the HudiJob script to delete the record from the Hudi table. A full delete record from AWS DMS for the human_resources.employee_details table looks like the following:
{ "Op": "D", "update_ts_dms": "2020-10-25 07:57:48.589284", "emp_no": 3, "name": "Jeff", "department": "Finance", "city": "Tokyo", "salary": 55000, "schema_name": "human_resources", "table_name": "employee_details"}
The schema_name, and table_name columns are added by AWS DMS because of the task configuration shared previously.update_ts_dms has been set as the value for TimestampColumnName S3 setting in AWS DMS S3 Endpoint.Op is added by AWS DMS for cdc and it indicates source DB operations in migrated S3 data.
We also set spark.serializer in the script. This setting is required for Hudi.
In HudiJob script, you can also find a few Python dict that store various Hudi configuration properties. These configurations are just for demo purposes; you have to adjust them based on your workload. For more information about Hudi configurations, see Configurations.
HudiJob is scheduled to run every 5 minutes by default. The frequency is set by the ScheduleToRunGlueJob parameter of the CloudFormation template. Make sure that you successfully run HudiJob at least one time after the source data lands in the raw S3 bucket. The screenshot in Step 6 of Running the initial load script section confirms that AWS DMS put the LOAD00000001.parquet file in the raw bucket at 11:54:41 AM and following screenshot confirms that the job execution started at 11:55 AM.

The job creates a Hudi table in the AWS Glue Data Catalog (see the following screenshot). The table is partitioned on the department column.

Granting AWS Lake Formation permissions
If you have AWS Lake Formation enabled, make sure that you grant Select permission on the human_resources.employee_details table to the role/user used to run Athena query. Similarly, you also have to grant Select permission on the human_resources.employee_details table to the LakeHouseRedshiftGlueAccessRole role so you can query human_resources.employee_details in Amazon Redshift.

Grant Drop permission on the human_resources database to LakeHouseExecuteLambdaFnsRole so that the template can delete the database when you delete the template. Also, the CloudFormation template does not roll back any AWS Lake Formation grants or changes that are manually applied.
Granting access to KMS key
The curated S3 bucket is encrypted by lakehouse-key, which is an AWS Key Management Service (AWS KMS) customer managed key created by AWS CloudFormation template.
To run the query in Athena, you have to add the ARN of the role/user used to run the Athena query in the Allow use of the key section in the key policy.


This will ensure that you don’t get com.amazonaws.services.s3.model.AmazonS3Exception: Access Denied (Service: Amazon S3; Status Code: 403; Error Code: AccessDenied; error while running your Athena query.
You might not have to execute the above KMS policy change if you have kept the default of granting access to the AWS account and the role/user used to run Athena query has the necessary KMS related policies attached to it.
Confirming job completion
When HudiJob is complete, you can see the files in the curated bucket.
- On the Resources tab, search for CuratedS3Bucket.
- Choose the Physical ID link.

The following screenshot shows the timestamp on the initial load.

- Navigate to the department=Finance prefix and select the Parquet file.
- Choose Select from.

- For File format, select Parquet.
- Choose Show file preview.
You can see the value of the timestamp in the update_ts_dms column.

Querying the Hudi table
You can now query your data in Amazon Athena or Amazon Redshift.
Querying in Amazon Athena
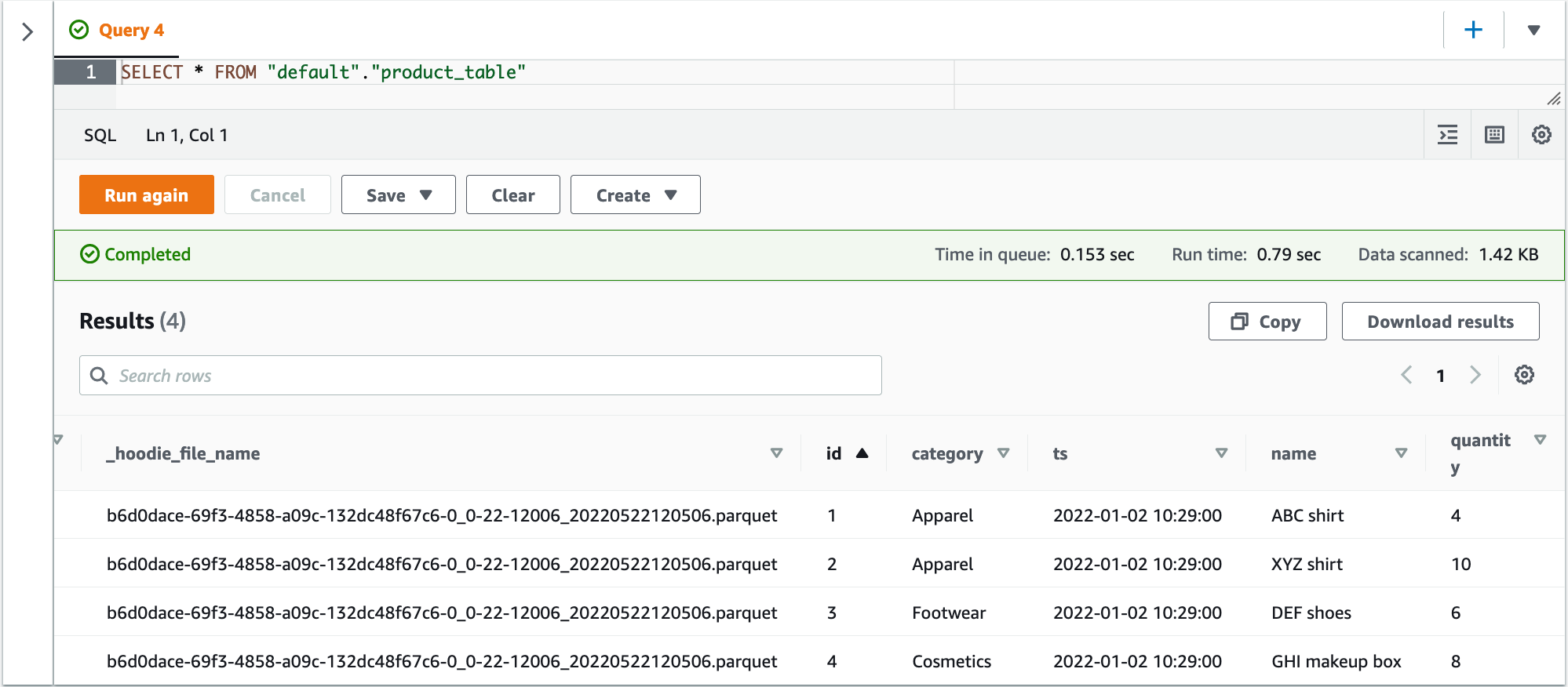
Query the human_resources.employee_details table in Amazon Athena with the following code:
SELECT emp_no,
name,
city,
salary,
department,
from_unixtime(update_ts_dms/1000000,'America/Los_Angeles') update_ts_dms_LA,
from_unixtime(update_ts_dms/1000000,'UTC') update_ts_dms_UTC
FROM "human_resources"."employee_details"
ORDER BY emp_no
The timestamp for all the records matches the timestamp in the update_ts_dms column in the earlier screenshot.

Querying in Redshift Spectrum
Read query your table in Redshift Spectrum for Apache Hudi support in Amazon Redshift.
- On the Amazon Redshift console, locate
lakehouse-redshift-cluster.
- Choose Query cluster.

- For Database name, enter
lakehouse_dw.
- For Database user, enter
rs_admin.
- For Database password, enter the password that you used for the
RedshiftDWMasterUserPassword parameter in the CloudFormation template.

- Enter the following query for the
human_resources.employee_details table:
SELECT emp_no,
name,
city,
salary,
department,
(TIMESTAMP 'epoch' + update_ts_dms/1000000 * interval '1 second') AT TIME ZONE 'utc' AT TIME ZONE 'america/los_angeles' update_ts_dms_LA,
(TIMESTAMP 'epoch' + update_ts_dms/1000000 * interval '1 second') AT TIME ZONE 'utc' update_ts_dms_UTC
FROM human_resources.employee_details
ORDER BY emp_no
The following screenshot shows the query output.

Running the incremental load script
We now run the IncrementalUpdatesAndInserts_TestStep2.sql script. The output shows that 6 statements were run.

AWS DMS now shows that it has replicated the new incremental changes. The changes are replicated at a frequency set in DMSBatchUnloadIntervalInSecs parameter of the CloudFormation stack.

This creates another Parquet file in the raw S3 bucket.

The incremental updates are loaded into the Hudi table according to the chosen frequency to run the job (the ScheduleToRunGlueJob parameter). The HudiJobscript uses job bookmarks to find out the incremental load so it only processes the new files brought in through AWS DMS.
Confirming job completion
Make sure that HudiJob runs successfully at least one time after the incremental file arrives in the raw bucket. The previous screenshot shows that the incremental file arrived in the raw bucket at 1:18:38 PM and the following screenshot shows that the job started at 1:20 PM.

Querying the changed data
You can now check the table in Athena and Amazon Redshift. Both results show that emp_no 3 is deleted, 8 and 9 have been added, and 2 and 5 have been updated.
The following screenshot shows the results in Athena.

The following screenshot shows the results in Redshift Spectrum.

AWS Glue Job HudiMoRCompactionJob
The CloudFormation template also deploys the AWS Glue job HudiMoRCompactionJob. This job is not scheduled; you only use it if you choose the MoR storage type. To execute the pipe for MoR storage type instead of CoW storage type, delete the CloudFormation stack and create it again. After creation, replace CoW in lakehouse-hudi-storage-type AWS Systems Manager Parameter with MoR.
If you use MoR storage type, the incremental updates are stored in log files. You can’t see the updates in the _ro (read optimized) view, but can see them in the _rt view. Amazon Athena documentation and Amazon Redshift documentation gives more details about support and considerations for Apache Hudi.
To see the incremental data in the _ro view, run the HudiMoRCompactionJob job. For more information about Hudi storage types and views, see Hudi Dataset Storage Types and Storage Types & Views. The following code is an example of the CLI command used to run HudiMoRCompactionJob job:
aws glue start-job-run --job-name HudiMoRCompactionJob --arguments="--DB_NAME=human_resources","--TABLE_NAME=employee_details","--IS_PARTITIONED=true"
You can decide on the frequency of running this job. You don’t have to run the job immediately after the HudiJob. You should run this job when you want the data to be available in the _ro view. You have to pass the schema name and the table name to this script so it knows the table to compact.
Additional considerations
The JAR file we use in this post has not been tested for AWS Glue streaming jobs. Additionally, there are some hardcoded Hudi options in the HudiJob script. These options are set for the sample table that we create for this post. Update the options based on your workload.
Conclusion
In this post, we created AWS Glue 2.0 jobs that moved the source upserts and deletes into Hudi tables. The code creates tables in the AWS GLue Data Catalog and updates partitions so you don’t have to run the crawlers to update them.
This post simplified your LakeHouse code base by giving you the benefits of Apache Hudi along with serverless AWS Glue. We also showed how to create an source to LakeHouse replication system using AWS Glue, AWS DMS, and Amazon Redshift with minimum overhead.
Appendix
We can write to Hudi tables because of the hudi-spark.jar file that we downloaded to our DependentJarsAndTempS3Bucket S3 bucket with the CloudFormation template. The path to this file is added as a dependency in both the AWS Glue jobs. This file is based on open-source Hudi. To create the JAR file, complete the following steps:
- Get Hudi 0.5.3 and unzip it using the following code:
wget https://github.com/apache/hudi/archive/release-0.5.3.zip
unzip hudi-release-0.5.3.zip
- Edit
Hudi pom.xml:
vi hudi-release-0.5.3/pom.xml
- Remove the following code to make the build process faster:
<module>packaging/hudi-hadoop-mr-bundle</module>
<module>packaging/hudi-hive-bundle</module>
<module>packaging/hudi-presto-bundle</module>
<module>packaging/hudi-utilities-bundle</module>
<module>packaging/hudi-timeline-server-bundle</module>
<module>docker/hoodie/hadoop</module>
<module>hudi-integ-test</module>
- Change the versions of all three dependencies of httpcomponents to 4.4.1. The following is the original code:
<!-- Httpcomponents -->
<dependency>
<groupId>org.apache.httpcomponents</groupId>
<artifactId>fluent-hc</artifactId>
<version>4.3.2</version>
</dependency>
<dependency>
<groupId>org.apache.httpcomponents</groupId>
<artifactId>httpcore</artifactId>
<version>4.3.2</version>
</dependency>
<dependency>
<groupId>org.apache.httpcomponents</groupId>
<artifactId>httpclient</artifactId>
<version>4.3.6</version>
</dependency>
The following is the replacement code:
<!-- Httpcomponents -->
<dependency>
<groupId>org.apache.httpcomponents</groupId>
<artifactId>fluent-hc</artifactId>
<version>4.4.1</version>
</dependency>
<dependency>
<groupId>org.apache.httpcomponents</groupId>
<artifactId>httpcore</artifactId>
<version>4.4.1</version>
</dependency>
<dependency>
<groupId>org.apache.httpcomponents</groupId>
<artifactId>httpclient</artifactId>
<version>4.4.1</version>
</dependency>
- Build the JAR file:
mvn clean package -DskipTests -DskipITs -f <Full path of the hudi-release-0.5.3 dir>
- You can now get the JAR from the following location:
hudi-release-0.5.3/packaging/hudi-spark-bundle/target/hudi-spark-bundle_2.11-0.5.3-rc2.jar
The other JAR dependency used in the AWS Glue jobs is spark-avro_2.11-2.4.4.jar.
About the Author
 Vishal Pathak is a Data Lab Solutions Architect at AWS. Vishal works with the customers on their use cases, architects a solution to solve their business problems and helps the customers build an scalable prototype. Prior to his journey in AWS, Vishal helped customers implement BI, DW and DataLake projects in US and Australia.
Vishal Pathak is a Data Lab Solutions Architect at AWS. Vishal works with the customers on their use cases, architects a solution to solve their business problems and helps the customers build an scalable prototype. Prior to his journey in AWS, Vishal helped customers implement BI, DW and DataLake projects in US and Australia.



 When the subscription is complete, you see the Effective date populated next to the product.
When the subscription is complete, you see the Effective date populated next to the product.







 Vishal Pathak is a Data Lab Solutions Architect at AWS. Vishal works with customers on their use cases, architects solutions to solve their business problems, and helps them build scalable prototypes. Prior to his journey in AWS, Vishal helped customers implement business intelligence, data warehouse, and data lake projects in the US and Australia.
Vishal Pathak is a Data Lab Solutions Architect at AWS. Vishal works with customers on their use cases, architects solutions to solve their business problems, and helps them build scalable prototypes. Prior to his journey in AWS, Vishal helped customers implement business intelligence, data warehouse, and data lake projects in the US and Australia. Noritaka Sekiyama is a Principal Big Data Architect on the AWS Glue team. He enjoys learning different use cases from customers and sharing knowledge about big data technologies with the wider community.
Noritaka Sekiyama is a Principal Big Data Architect on the AWS Glue team. He enjoys learning different use cases from customers and sharing knowledge about big data technologies with the wider community. Anand Prakash is a Senior Solutions Architect at AWS Data Lab. Anand focuses on helping customers design and build AI/ML, data analytics, and database solutions to accelerate their path to production.
Anand Prakash is a Senior Solutions Architect at AWS Data Lab. Anand focuses on helping customers design and build AI/ML, data analytics, and database solutions to accelerate their path to production.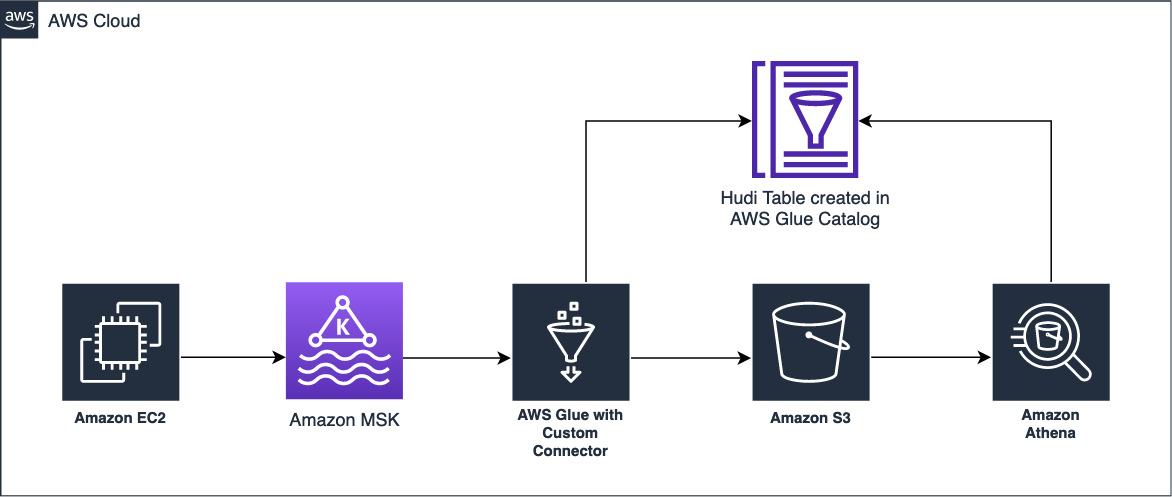

























 Vishal Pathak is a Data Lab Solutions Architect at AWS. Vishal works with the customers on their use cases, architects a solution to solve their business problems and helps the customers build an scalable prototype. Prior to his journey in AWS, Vishal helped customers implement BI, DW and DataLake projects in US and Australia.
Vishal Pathak is a Data Lab Solutions Architect at AWS. Vishal works with the customers on their use cases, architects a solution to solve their business problems and helps the customers build an scalable prototype. Prior to his journey in AWS, Vishal helped customers implement BI, DW and DataLake projects in US and Australia.
































































