Post Syndicated from Ravi Itha original https://aws.amazon.com/blogs/big-data/simplify-operational-data-processing-in-data-lakes-using-aws-glue-and-apache-hudi/
The Analytics specialty practice of AWS Professional Services (AWS ProServe) helps customers across the globe with modern data architecture implementations on the AWS Cloud. A modern data architecture is an evolutionary architecture pattern designed to integrate a data lake, data warehouse, and purpose-built stores with a unified governance model. It focuses on defining standards and patterns to integrate data producers and consumers and move data between data lakes and purpose-built data stores securely and efficiently. Out of the many data producer systems that feed data to a data lake, operational databases are most prevalent, where operational data is stored, transformed, analyzed, and finally used to enhance business operations of an organization. With the emergence of open storage formats such as Apache Hudi and its native support from AWS Glue for Apache Spark, many AWS customers have started adding transactional and incremental data processing capabilities to their data lakes.
AWS has invested in native service integration with Apache Hudi and published technical contents to enable you to use Apache Hudi with AWS Glue (for example, refer to Introducing native support for Apache Hudi, Delta Lake, and Apache Iceberg on AWS Glue for Apache Spark, Part 1: Getting Started). In AWS ProServe-led customer engagements, the use cases we work on usually come with technical complexity and scalability requirements. In this post, we discuss a common use case in relation to operational data processing and the solution we built using Apache Hudi and AWS Glue.
Use case overview
AnyCompany Travel and Hospitality wanted to build a data processing framework to seamlessly ingest and process data coming from operational databases (used by reservation and booking systems) in a data lake before applying machine learning (ML) techniques to provide a personalized experience to its users. Due to the sheer volume of direct and indirect sales channels the company has, its booking and promotions data are organized in hundreds of operational databases with thousands of tables. Of those tables, some are larger (such as in terms of record volume) than others, and some are updated more frequently than others. In the data lake, the data to be organized in the following storage zones:
- Source-aligned datasets – These have an identical structure to their counterparts at the source
- Aggregated datasets – These datasets are created based on one or more source-aligned datasets
- Consumer-aligned datasets – These are derived from a combination of source-aligned, aggregated, and reference datasets enriched with relevant business and transformation logics, usually fed as inputs to ML pipelines or any consumer applications
The following are the data ingestion and processing requirements:
- Replicate data from operational databases to the data lake, including insert, update, and delete operations.
- Keep the source-aligned datasets up to date (typically within the range of 10 minutes to a day) in relation to their counterparts in the operational databases, ensuring analytics pipelines refresh consumer-aligned datasets for downstream ML pipelines in a timely fashion. Moreover, the framework should consume compute resources as optimally as possible per the size of the operational tables.
- To minimize DevOps and operational overhead, the company wanted to templatize the source code wherever possible. For example, to create source-aligned datasets in the data lake for 3,000 operational tables, the company didn’t want to deploy 3,000 separate data processing jobs. The smaller the number of jobs and scripts, the better.
- The company wanted the ability to continue processing operational data in the secondary Region in the rare event of primary Region failure.
As you can guess, the Apache Hudi framework can solve the first requirement. Therefore, we will put our emphasis on the other requirements. We begin with a Data lake reference architecture followed by an overview of operational data processing framework. By showing you our open-source solution on GitHub, we delve into framework components and walk through their design and implementation aspects. Finally, by testing the framework, we summarize how it meets the aforementioned requirements.
Data lake reference architecture
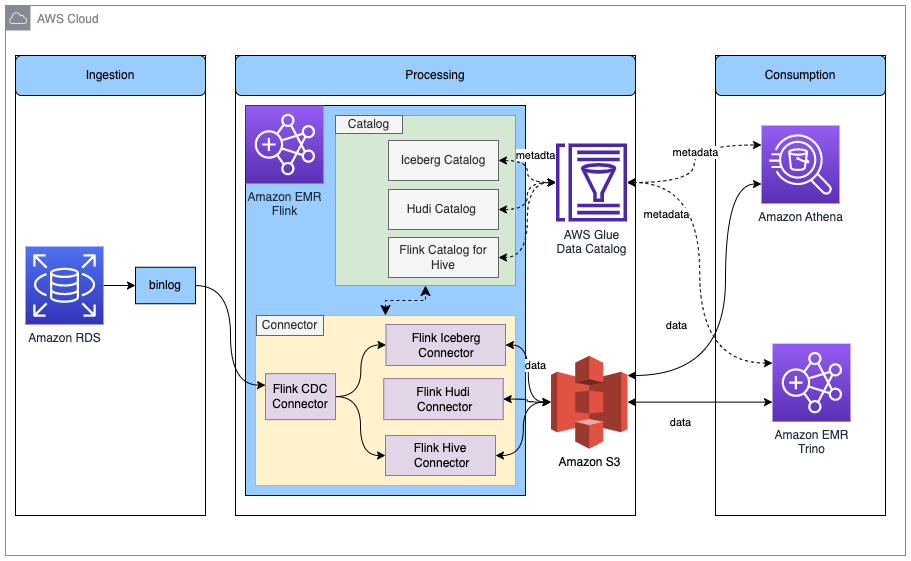
Let’s begin with a big picture: a data lake solves a variety of analytics and ML use cases dealing with internal and external data producers and consumers. The following diagram represents a generic data lake architecture. To ingest data from operational databases to an Amazon Simple Storage Service (Amazon S3) staging bucket of the data lake, either AWS Database Migration Service (AWS DMS) or any AWS partner solution from AWS Marketplace that has support for change data capture (CDC) can fulfill the requirement. AWS Glue is used to create source-aligned and consumer-aligned datasets and separate AWS Glue jobs to do feature engineering part of ML engineering and operations. Amazon Athena is used for interactive querying and AWS Lake Formation is used for access controls.

Operational data processing framework
The operational data processing (ODP) framework contains three components: File Manager, File Processor, and Configuration Manager. Each component runs independently to solve a portion of the operational data processing use case. We have open-sourced this framework on GitHub—you can clone the code repo and inspect it while we walk you through the design and implementation of the framework components. The source code is organized in three folders, one for each component, and if you customize and adopt this framework for your use case, we recommend promoting these folders as separate code repositories in your version control system. Consider using the following repository names:
aws-glue-hudi-odp-framework-file-manageraws-glue-hudi-odp-framework-file-processoraws-glue-hudi-odp-framework-config-manager
With this modular approach, you can independently deploy the components to your data lake environment by following your preferred CI/CD processes. As illustrated in the preceding diagram, these components are deployed in conjunction with a CDC solution.
Component 1: File Manager
File Manager detects files emitted by a CDC process such as AWS DMS and tracks them in an Amazon DynamoDB table. As shown in the following diagram, it consists of an Amazon EventBridge event rule, an Amazon Simple Queue Service (Amazon SQS) queue, an AWS Lambda function, and a DynamoDB table. The EventBridge rule uses Amazon S3 Event Notifications to detect the arrival of CDC files in the S3 bucket. The event rule forwards the object event notifications to the SQS queue as messages. The File Manager Lambda function consumes those messages, parses the metadata, and inserts the metadata to the DynamoDB table odpf_file_tracker. These records will then be processed by File Processor, which we discuss in the next section.

Component 2: File Processor
File Processor is the workhorse of the ODP framework. It processes files from the S3 staging bucket, creates source-aligned datasets in the raw S3 bucket, and adds or updates metadata for the datasets (AWS Glue tables) in the AWS Glue Data Catalog.
We use the following terminology when discussing File Processor:
- Refresh cadence – This represents the data ingestion frequency (for example, 10 minutes). It usually goes with AWS Glue worker type (one of G.1X, G.2X, G.4X, G.8X, G.025X, and so on) and batch size.
- Table configuration – This includes the Hudi configuration (primary key, partition key, pre-combined key, and table type (Copy on Write or Merge on Read)), table data storage mode (historical or current snapshot), S3 bucket used to store source-aligned datasets, AWS Glue database name, AWS Glue table name, and refresh cadence.
- Batch size – This numeric value is used to split tables into smaller batches and process their respective CDC files in parallel. For example, a configuration of 50 tables with a 10-minute refresh cadence and a batch size of 5 results in a total of 10 AWS Glue job runs, each processing CDC files for 5 tables.
- Table data storage mode – There are two options:
- Historical – This table in the data lake stores historical updates to records (always append).
- Current snapshot – This table in the data lake stores latest versioned records (upserts) with the ability to use Hudi time travel for historical updates.
- File processing state machine – It processes CDC files that belong to tables that share a common refresh cadence.
- EventBridge rule association with the file processing state machine – We use a dedicated EventBridge rule for each refresh cadence with the file processing state machine as target.
- File processing AWS Glue job – This is a configuration-driven AWS Glue extract, transform, and load (ETL) job that processes CDC files for one or more tables.
File Processor is implemented as a state machine using AWS Step Functions. Let’s use an example to understand this. The following diagram illustrates running File Processor state machine with a configuration that includes 18 operational tables, a refresh cadence of 10 minutes, a batch size of 5, and an AWS Glue worker type of G.1X.

The workflow includes the following steps:
- The EventBridge rule triggers the File Processor state machine every 10 minutes.
- Being the first state in the state machine, the Batch Manager Lambda function reads configurations from DynamoDB tables.
- The Lambda function creates four batches: three of them will be mapped to five operational tables each, and the fourth one is mapped to three operational tables. Then it feeds the batches to the Step Functions Map state.
- For each item in the Map state, the File Processor Trigger Lambda function will be invoked, which in turn runs the File Processor AWS Glue job.
- Each AWS Glue job performs the following actions:
- Checks the status of an operational table and acquires a lock when it is not processed by any other job. The
odpf_file_processing_trackerDynamoDB table is used for this purpose. When a lock is acquired, it inserts a record in the DynamoDB table with the statusupdating_tablefor the first time; otherwise, it updates the record. - Processes the CDC files for the given operational table from the S3 staging bucket and creates a source-aligned dataset in the S3 raw bucket. It also updates technical metadata in the AWS Glue Data Catalog.
- Updates the status of the operational table to
completedin theodpf_file_processing_trackertable. In case of processing errors, it updates the status to refresh_error and logs the stack trace. - It also inserts this record into the
odpf_file_processing_tracker_historyDynamoDB table along with additional details such as insert, update, and delete row counts. - Moves the records that belong to successfully processed CDC files from
odpf_file_trackerto theodpf_file_tracker_historytable withfile_ingestion_statusset toraw_file_processed. - Moves to the next operational table in the given batch.
- Note: a failure to process CDC files for one of the operational tables of a given batch does not impact the processing of other operational tables.
- Checks the status of an operational table and acquires a lock when it is not processed by any other job. The
Component 3: Configuration Manager
Configuration Manager is used to insert configuration details to the odpf_batch_config and odpf_raw_table_config tables. To keep this post concise, we provide two architecture patterns in the code repo and leave the implementation details to you.
Solution overview
Let’s test the ODP framework by replicating data from 18 operational tables to a data lake and creating source-aligned datasets with 10-minute refresh cadence. We use Amazon Relational Database Service (Amazon RDS) for MySQL to set up an operational database with 18 tables, upload the New York City Taxi – Yellow Trip Data dataset, set up AWS DMS to replicate data to Amazon S3, process the files using the framework, and finally validate the data using Amazon Athena.
Create S3 buckets
For instructions on creating an S3 bucket, refer to Creating a bucket. For this post, we create the following buckets:
odpf-demo-staging-EXAMPLE-BUCKET– You will use this to migrate operational data using AWS DMSodpf-demo-raw-EXAMPLE-BUCKET– You will use this to store source-aligned datasetsodpf-demo-code-artifacts-EXAMPLE-BUCKET– You will use this to store code artifacts
Deploy File Manager and File Processor
Deploy File Manager and File Processor by following instructions from this README and this README, respectively.
Set up Amazon RDS for MySQL
Complete the following steps to set up Amazon RDS for MySQL as the operational data source:
- Provision Amazon RDS for MySQL. For instructions, refer to Create and Connect to a MySQL Database with Amazon RDS.
- Connect to the database instance using MySQL Workbench or DBeaver.
- Create a database (schema) by running the SQL command
CREATE DATABASE taxi_trips;. - Create 18 tables by running the SQL commands in the ops_table_sample_ddl.sql script.
Populate data to the operational data source
Complete the following steps to populate data to the operational data source:
- To download the New York City Taxi – Yellow Trip Data dataset for January 2021 (Parquet file), navigate to NYC TLC Trip Record Data, expand 2021, and choose Yellow Taxi Trip records. A file called
yellow_tripdata_2021-01.parquetwill be downloaded to your computer. - On the Amazon S3 console, open the bucket
odpf-demo-staging-EXAMPLE-BUCKETand create a folder callednyc_yellow_trip_data. - Upload the
yellow_tripdata_2021-01.parquetfile to the folder. - Navigate to the bucket
odpf-demo-code-artifacts-EXAMPLE-BUCKETand create a folder calledglue_scripts. - Download the file load_nyc_taxi_data_to_rds_mysql.py from the GitHub repo and upload it to the folder.
- Create an AWS Identity and Access Management (IAM) policy called
load_nyc_taxi_data_to_rds_mysql_s3_policy. For instructions, refer to Creating policies using the JSON editor. Use the odpf_setup_test_data_glue_job_s3_policy.json policy definition. - Create an IAM role called
load_nyc_taxi_data_to_rds_mysql_glue_role. Attach the policy created in the previous step. - On the AWS Glue console, create a connection for Amazon RDS for MySQL. For instructions, refer to Adding a JDBC connection using your own JDBC drivers and Setting up a VPC to connect to Amazon RDS data stores over JDBC for AWS Glue. Name the connection as
odpf_demo_rds_connection. - In the navigation pane of the AWS Glue console, choose Glue ETL jobs, Python Shell script editor, and Upload and edit an existing script under Options.
- Choose the file
load_nyc_taxi_data_to_rds_mysql.pyand choose Create. - Complete the following steps to create your job:
- Provide a name for the job, such as
load_nyc_taxi_data_to_rds_mysql. - For IAM role, choose
load_nyc_taxi_data_to_rds_mysql_glue_role. - Set Data processing units to
1/16 DPU. - Under Advanced properties, Connections, select the connection you created earlier.
- Under Job parameters, add the following parameters:
input_sample_data_path=s3://odpf-demo-staging-EXAMPLE-BUCKET/nyc_yellow_trip_data/yellow_tripdata_2021-01.parquetschema_name=taxi_tripstable_name=table_1rds_connection_name=odpf_demo_rds_connection
- Choose Save.
- Provide a name for the job, such as
- On the Actions menu, run the job.
- Go back to your MySQL Workbench or DBeaver and validate the record count by running the SQL command
select count(1) row_count from taxi_trips.table_1. You will get an output of1369769. - Populate the remaining 17 tables by running the SQL commands from the populate_17_ops_tables_rds_mysql.sql script.
- Get the row count from the 18 tables by running the SQL commands from the ops_data_validation_query_rds_mysql.sql script. The following screenshot shows the output.

Configure DynamoDB tables
Complete the following steps to configure the DynamoDB tables:
- Download file load_ops_table_configs_to_ddb.py from the GitHub repo and upload it to the folder
glue_scriptsin the S3 bucketodpf-demo-code-artifacts-EXAMPLE-BUCKET. - Create an IAM policy called
load_ops_table_configs_to_ddb_ddb_policy. Use the odpf_setup_test_data_glue_job_ddb_policy.json policy definition. - Create an IAM role called
load_ops_table_configs_to_ddb_glue_role. Attach the policy created in the previous step. - On the AWS Glue console, choose Glue ETL jobs, Python Shell script editor, and Upload and edit an existing script under Options.
- Choose the file
load_ops_table_configs_to_ddb.pyand choose Create. - Complete the following steps to create a job:
- Provide a name, such as
load_ops_table_configs_to_ddb. - For IAM role, choose
load_ops_table_configs_to_ddb_glue_role. - Set Data processing units to
1/16 DPU. - Under Job parameters, add the following parameters
batch_config_ddb_table_name=odpf_batch_configraw_table_config_ddb_table_name=odpf_demo_taxi_trips_rawaws_region= e.g.,us-west-1
- Choose Save.
- Provide a name, such as
- On the Actions menu, run the job.
- On the DynamoDB console, get the item count from the tables. You will find 1 item in the
odpf_batch_configtable and 18 items in theodpf_demo_taxi_trips_rawtable.
Set up a database in AWS Glue
Complete the following steps to create a database:
- On the AWS Glue console, under Data catalog in the navigation pane, choose Databases.
- Create a database called
odpf_demo_taxi_trips_raw.
Set up AWS DMS for CDC
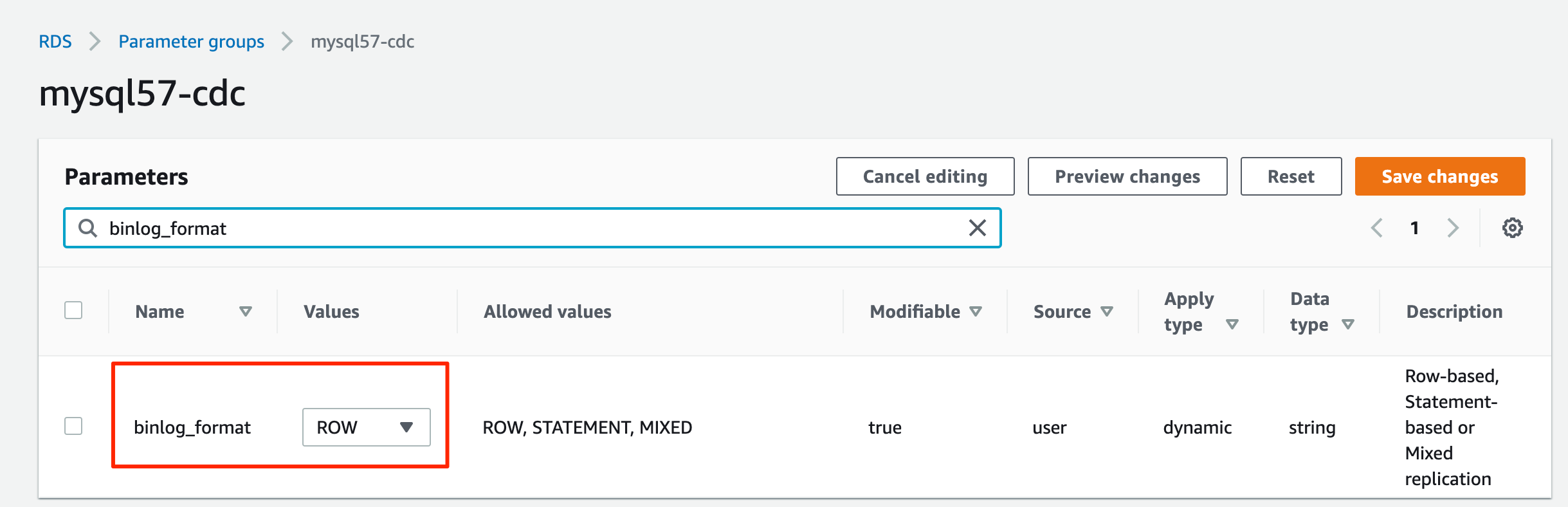
Complete the following steps to set up AWS DMS for CDC:
- Create an AWS DMS replication instance. For Instance class, choose dms.t3.medium.
- Create a source endpoint for Amazon RDS for MySQL.
- Create target endpoint for Amazon S3. To configure the S3 endpoint settings, use the JSON definition from dms_s3_endpoint_setting.json.
- Create an AWS DMS task.
- Use the source and target endpoints created in the previous steps.
- To create AWS DMS task mapping rules, use the JSON definition from dms_task_mapping_rules.json.
- Under Migration task startup configuration, select Automatically on create.
- When the AWS DMS task starts running, you will see a task summary similar to the following screenshot.

- In the Table statistics section, you will see an output similar to the following screenshot. Here, the Full load rows and Total rows columns are important metrics whose counts should match with the record volumes of the 18 tables in the operational data source.

- As a result of successful full load completion, you will find Parquet files in the S3 staging bucket—one Parquet file per table in a dedicated folder, similar to the following screenshot. Similarly, you will find 17 such folders in the bucket.

File Manager output
The File Manager Lambda function consumes messages from the SQS queue, extracts metadata for the CDC files, and inserts one item per file to the odpf_file_tracker DynamoDB table. When you check the items, you will find 18 items with file_ingestion_status set to raw_file_landed, as shown in the following screenshot.
![]()
File Processor output
- On the subsequent tenth minute (since the activation of the EventBridge rule), the event rule triggers the File Processor state machine. On the Step Functions console, you will notice that the state machine is invoked, as shown in the following screenshot.

- As shown in the following screenshot, the Batch Generator Lambda function creates four batches and constructs a Map state for parallel running of the File Processor Trigger Lambda function.

- Then, the File Processor Trigger Lambda function runs the File Processor Glue Job, as shown in the following screenshot.

- Then, you will notice that the File Processor Glue Job runs create source-aligned datasets in Hudi format in the S3 raw bucket. For Table 1, you will see an output similar to the following screenshot. There will be 17 such folders in the S3 raw bucket.

- Finally, in AWS Glue Data Catalog, you will notice 18 tables created in the
odpf_demo_taxi_trips_rawdatabase, similar to the following screenshot.

Data validation
Complete the following steps to validate the data:
- On the Amazon Athena console, open the query editor, and select a workgroup or create a new workgroup.
- Choose
AwsDataCatalogfor Data source andodpf_demo_taxi_trips_rawfor Database. - Run the raw_data_validation_query_athena.sql SQL query. You will get an output similar to the following screenshot.

Validation summary: The counts in Amazon Athena match with the counts of the operational tables and it proves that the ODP framework has processed all the files and records successfully. This concludes the demo. To test additional scenarios, refer to Extended Testing in the code repo.
Outcomes
Let’s review how the ODP framework addressed the aforementioned requirements.
- As discussed earlier in this post, by logically grouping tables by refresh cadence and associating them to EventBridge rules, we ensured that the source-aligned tables are refreshed by the File Processor AWS Glue jobs. With the AWS Glue worker type configuration setting, we selected the appropriate compute resources while running the AWS Glue jobs (the instances of the AWS Glue job).
- By applying table-specific configurations (from
odpf_batch_configandodpf_raw_table_config) dynamically, we were able to use one AWS Glue job to process CDC files for 18 tables. - You can use this framework to support a variety of data migration use cases that require quicker data migration from on-premises storage systems to data lakes or analytics platforms on AWS. You can reuse File Manager as is and customize File Processor to work with other storage frameworks such as Apache Iceberg, Delta Lake, and purpose-built data stores such as Amazon Aurora and Amazon Redshift.
- To understand how the ODP framework met the company’s disaster recovery (DR) design criterion, we first need to understand the DR architecture strategy at a high level. The DR architecture strategy has the following aspects:
- One AWS account and two AWS Regions are used for primary and secondary environments.
- The data lake infrastructure in the secondary Region is kept in sync with the one in the primary Region.
- Data is stored in S3 buckets, metadata data is stored in the AWS Glue Data Catalog, and access controls in Lake Formation are replicated from the primary to secondary Region.
- The data lake source and target systems have their respective DR environments.
- CI/CD tooling (version control, CI server, and so on) are to be made highly available.
- The DevOps team needs to be able to deploy CI/CD pipelines of analytics frameworks (such as this ODP framework) to either the primary or secondary Region.
- As you can imagine, disaster recovery on AWS is a vast subject, so we keep our discussion to the last design aspect.
By designing the ODP framework with three components and externalizing operational table configurations to DynamoDB global tables, the company was able to deploy the framework components to the secondary Region (in the rare event of a single-Region failure) and continue to process CDC files from the point it last processed in the primary Region. Because the CDC file tracking and processing audit data is replicated to the DynamoDB replica tables in the secondary Region, the File Manager microservice and File Processor can seamlessly run.
Clean up
When you’re finished testing this framework, you can delete the provisioned AWS resources to avoid any further charges.
Conclusion
In this post, we took a real-world operational data processing use case and presented you the framework we developed at AWS ProServe. We hope this post and the operational data processing framework using AWS Glue and Apache Hudi will expedite your journey in integrating operational databases into your modern data platforms built on AWS.
About the authors
 Ravi Itha is a Principal Consultant at AWS Professional Services with specialization in data and analytics and generalist background in application development. Ravi helps customers with enterprise data strategy initiatives across insurance, airlines, pharmaceutical, and financial services industries. In his 6-year tenure at Amazon, Ravi has helped the AWS builder community by publishing approximately 15 open-source solutions (accessible via GitHub handle), four blogs, and reference architectures. Outside of work, he is passionate about reading India Knowledge Systems and practicing Yoga Asanas.
Ravi Itha is a Principal Consultant at AWS Professional Services with specialization in data and analytics and generalist background in application development. Ravi helps customers with enterprise data strategy initiatives across insurance, airlines, pharmaceutical, and financial services industries. In his 6-year tenure at Amazon, Ravi has helped the AWS builder community by publishing approximately 15 open-source solutions (accessible via GitHub handle), four blogs, and reference architectures. Outside of work, he is passionate about reading India Knowledge Systems and practicing Yoga Asanas.
 Srinivas Kandi is a Data Architect at AWS Professional Services. He leads customer engagements related to data lakes, analytics, and data warehouse modernizations. He enjoys reading history and civilizations.
Srinivas Kandi is a Data Architect at AWS Professional Services. He leads customer engagements related to data lakes, analytics, and data warehouse modernizations. He enjoys reading history and civilizations.

































 Raj Ramasubbu is a Sr. Analytics Specialist Solutions Architect focused on big data and analytics and AI/ML with Amazon Web Services. He helps customers architect and build highly scalable, performant, and secure cloud-based solutions on AWS. Raj provided technical expertise and leadership in building data engineering, big data analytics, business intelligence, and data science solutions for over 18 years prior to joining AWS. He helped customers in various industry verticals like healthcare, medical devices, life science, retail, asset management, car insurance, residential REIT, agriculture, title insurance, supply chain, document management, and real estate.
Raj Ramasubbu is a Sr. Analytics Specialist Solutions Architect focused on big data and analytics and AI/ML with Amazon Web Services. He helps customers architect and build highly scalable, performant, and secure cloud-based solutions on AWS. Raj provided technical expertise and leadership in building data engineering, big data analytics, business intelligence, and data science solutions for over 18 years prior to joining AWS. He helped customers in various industry verticals like healthcare, medical devices, life science, retail, asset management, car insurance, residential REIT, agriculture, title insurance, supply chain, document management, and real estate. Rahul Sonawane is a Principal Analytics Solutions Architect at AWS with AI/ML and Analytics as his area of specialty.
Rahul Sonawane is a Principal Analytics Solutions Architect at AWS with AI/ML and Analytics as his area of specialty. Sundeep Kumar is a Sr. Data Architect, Data Lake at AWS, helping customers build data lake and analytics platform and solutions. When not building and designing data lakes, Sundeep enjoys listening music and playing guitar.
Sundeep Kumar is a Sr. Data Architect, Data Lake at AWS, helping customers build data lake and analytics platform and solutions. When not building and designing data lakes, Sundeep enjoys listening music and playing guitar.

















 Suthan Phillips is a Big Data Architect for Amazon EMR at AWS. He works with customers to provide best practice and technical guidance and helps them achieve highly scalable, reliable and secure solutions for complex applications on Amazon EMR. In his spare time, he enjoys hiking and exploring the Pacific Northwest.
Suthan Phillips is a Big Data Architect for Amazon EMR at AWS. He works with customers to provide best practice and technical guidance and helps them achieve highly scalable, reliable and secure solutions for complex applications on Amazon EMR. In his spare time, he enjoys hiking and exploring the Pacific Northwest. Dylan Qu is an AWS solutions architect responsible for providing architectural guidance across the full AWS stack with a focus on Data Analytics, AI/ML and DevOps.
Dylan Qu is an AWS solutions architect responsible for providing architectural guidance across the full AWS stack with a focus on Data Analytics, AI/ML and DevOps.