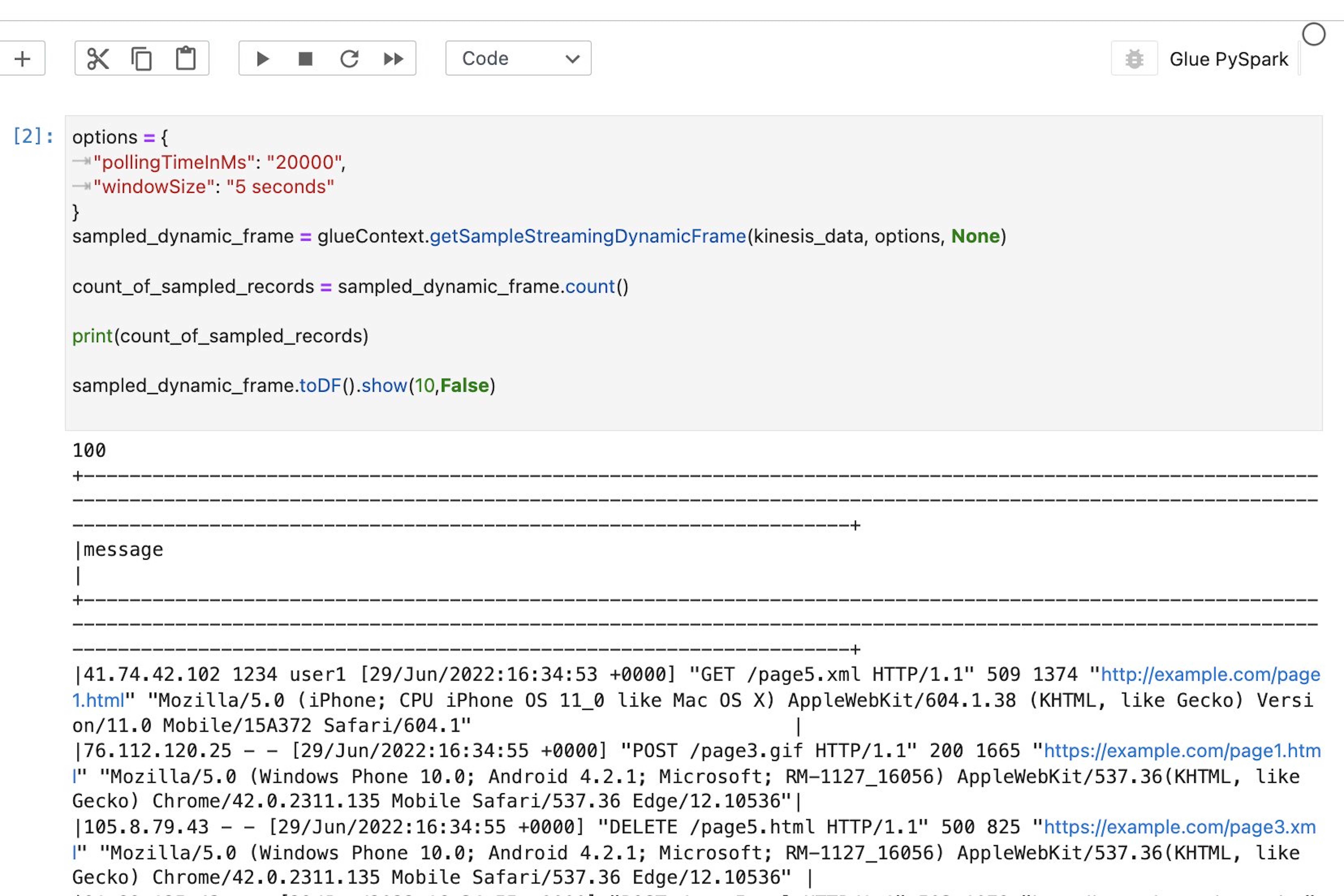
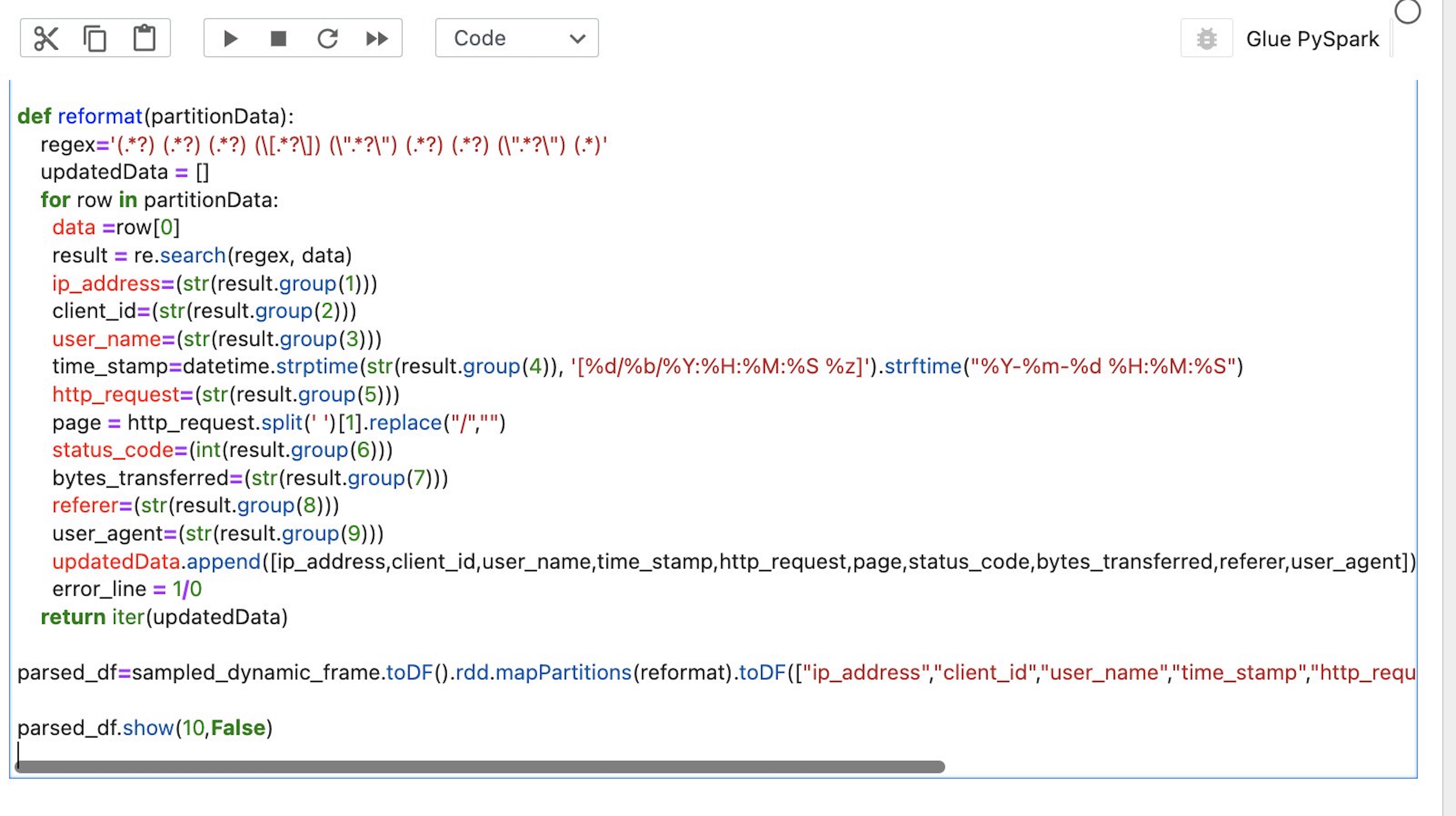
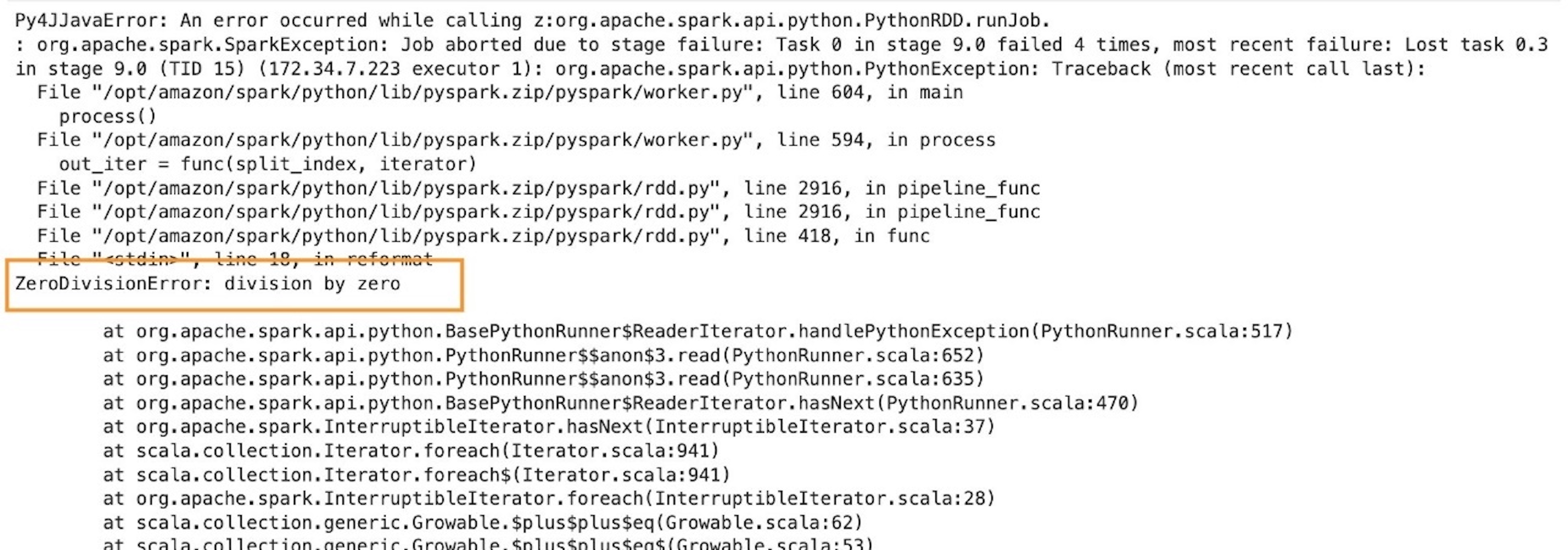
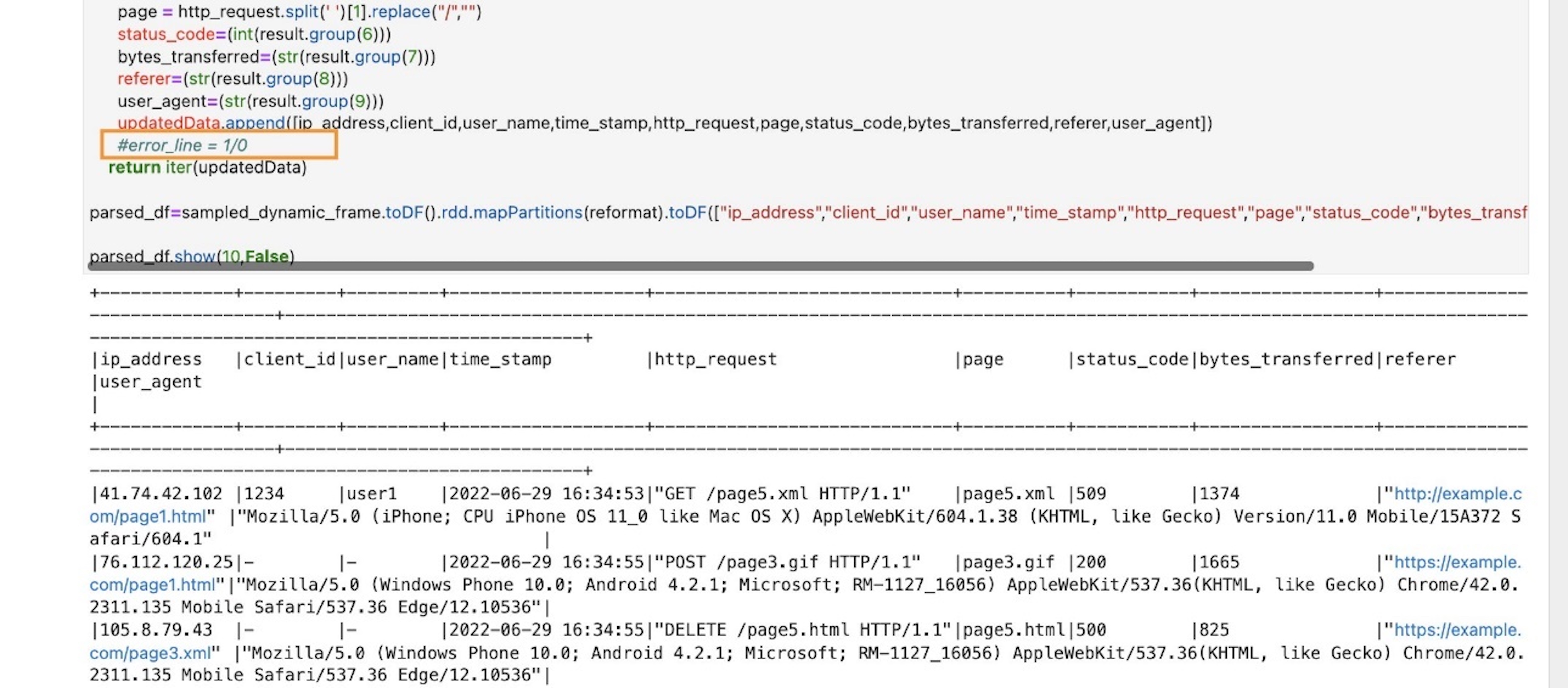
Post Syndicated from Jack Mazanec original https://aws.amazon.com/blogs/big-data/choose-the-k-nn-algorithm-for-your-billion-scale-use-case-with-opensearch/
When organizations set out to build machine learning (ML) applications such as natural language processing (NLP) systems, recommendation engines, or search-based systems, often times k-Nearest Neighbor (k-NN) search will be used at some point in the workflow. As the number of data points reaches the hundreds of millions or even billions, scaling a k-NN search system can be a major challenge. Applying Approximate Nearest Neighbor (ANN) search is a great way to overcome this challenge.
The k-NN problem is relatively simple compared to other ML techniques: given a set of points and a query, find the k nearest points in the set to the query. The naive solution is equally understandable: for each point in the set, compute its distance from the query and keep track of the top k along the way.

The problem with this naive approach is that it doesn’t scale particularly well. The runtime search complexity is O(Nlogk), where N is the number of vectors and k is the number of nearest neighbors. Although this may not be noticeable when the set contains thousands of points, it becomes noticeable when the size gets into the millions. Although some exact k-NN algorithms can speed search up, they tend to perform similarly to the naive approach in higher dimensions.
Enter ANN search. We can reduce the runtime search latency if we loosen a few constraints on the k-NN problem:
- Allow indexing to take longer
- Allow more space to be used at query time
- Allow the search to return an approximation of the k-NN in the set
Several different algorithms have been discovered to do just that.
OpenSearch is a community-driven, Apache 2.0-licensed, open-source search and analytics suite that makes it easy to ingest, search, visualize, and analyze data. The OpenSearch k-NN plugin provides the ability to use some of these algorithms within an OpenSearch cluster. In this post, we discuss the different algorithms that are supported and run experiments to see some of the trade-offs between them.
Hierarchical Navigable Small Worlds algorithm
The Hierarchical Navigable Small Worlds algorithm (HNSW) is one of the most popular algorithms out there for ANN search. It was the first algorithm that the k-NN plugin supported, using a very efficient implementation from the nmslib similarity search library. It has one of the best query latency vs. recall trade-offs and doesn’t require any training. The core idea of the algorithm is to build a graph with edges connecting index vectors that are close to each other. Then, on search, this graph is partially traversed to find the approximate nearest neighbors to the query vector. To steer the traversal towards the query’s nearest neighbors, the algorithm always visits the closest candidate to the query vector next.
But which vector should the traversal start from? It could just pick a random vector, but for a large index, this might be very far from the query’s actual nearest neighbors, leading to poor results. To pick a vector that is generally close to the query vector to start from, the algorithm builds not just one graph, but a hierarchy of graphs. All vectors are added to the bottom layer, and then a random subset of those are added to the layer above, and then a subset of those are added to the layer above that, and so on.
During search, we start from a random vector in the top layer, partially traverse the graph to find (approximately) the nearest point to the query vector in that layer, and then use this vector as the starting point for our traversal of the layer below. We repeat this until we get to the bottom layer. At the bottom layer, we perform the traversal, but this time, instead of just searching for the nearest neighbor, we keep track of the k-nearest neighbors that are visited along the way.
The following figure illustrates this process (inspired from the image in original paper Efficient and robust approximate nearest neighbor search using Hierarchical Navigable Small World graphs).

You can tune three parameters for HNSW:
- m – The maximum number of edges a vector will get in a graph. The higher this number is, the more memory the graph will consume, but the better the search approximation may be.
- ef_search – The size of the queue of the candidate nodes to visit during traversal. When a node is visited, its neighbors are added to the queue to be visited in the future. When this queue is empty, the traversal will end. A larger value will increase search latency, but may provide better search approximation.
- ef_construction – Very similar to
ef_search. When a node is to be inserted into the graph, the algorithm will find its m edges by querying the graph with the new node as the query vector. This parameter controls the candidate queue size for this traversal. A larger value will increase index latency, but may provide a better search approximation.
For more information on HNSW, you can read through the paper Efficient and robust approximate nearest neighbor search using Hierarchical Navigable Small World graphs.
Memory consumption
Although HNSW provides very good approximate nearest neighbor search at low latencies, it can consume a large amount of memory. Each HNSW graph uses roughly 1.1 * (4 * d + 8 * m) * num_vectors bytes of memory:
d is the dimension of the vectorsm is the algorithm parameter that controls the number of connections each node will have in a layernum_vectors is the number of vectors in the index
To ensure durability and availability, especially when running production workloads, OpenSearch indexes are recommended to have at least one replica shard. Therefore, the memory requirement is multiplied by (1 + number of replicas). For use cases where the data size is 1 billion vectors of 128 dimensions each and m is set to the default value of 16, the estimated amount of memory required would be:
1.1 * (4 * 128 + 8 * 16) * 1,000,000,000 * 2 = 1,408 GB.
If we increase the size of vectors to 512, for example, and the m to 100, which is recommended for vectors with high intrinsic dimensionality, some use cases can require a total memory of approximately 4 TB.
With OpenSearch, we can always horizontally scale the cluster to handle this memory requirement. However, this comes at the expense of raising infrastructure costs. For cases where scaling doesn’t make sense, options to reduce the memory footprint of the k-NN system need to be explored. Fortunately, there are algorithms that we can use to do this.
Inverted File System algorithm
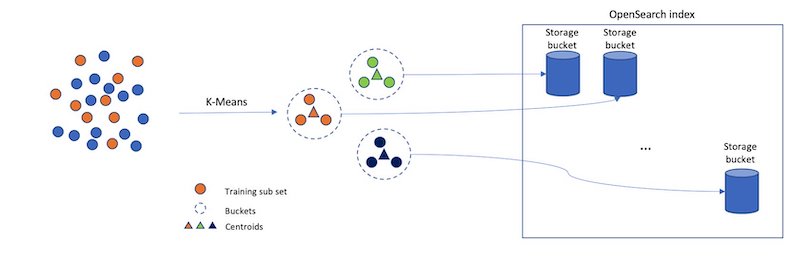
Consider a different approach for approximating a nearest neighbor search: separate your index vectors into a set of buckets, then, to reduce your search time, only search through a subset of these buckets. From a high level, this is what the Inverted File System (IVF) ANN algorithm does. In OpenSearch 1.2, the k-NN plugin introduced support for the implementation of IVF by Faiss. Faiss is an open-sourced library from Meta for efficient similarity search and clustering of dense vectors.
However, if we just randomly split up our vectors into different buckets, and only search a subset of them, this will be a poor approximation. The IVF algorithm uses a more elegant approach. First, before indexing begins, it assigns each bucket a representative vector. When a vector is indexed, it gets added to the bucket that has the closest representative vector. This way, vectors that are closer to each other are placed roughly in the same or nearby buckets.
To determine what the representative vectors for the buckets are, the IVF algorithm requires a training step. In this step, k-Means clustering is run on a set of training data, and the centroids it produces become the representative vectors. The following diagram illustrates this process.

IVF has two parameters:
- nlist – The number of buckets to create. More buckets will result in longer training times, but may improve the granularity of the search.
- nprobes – The number of buckets to search. This parameter is fairly straightforward. The more buckets that are searched, the longer the search will take, but the better the approximation.
Memory consumption
In general, IVF requires less memory than HNSW because IVF doesn’t need to store a set of edges for each indexed vector.
We estimate that IVF will roughly require the following amount of memory:
1.1 * (((4 * dimension) * num_vectors) + (4 * nlist * dimension)) bytes
For the case explored for HNSW where there are 1,000,000,000 128-dimensional vectors with one layer of replication, an IVF algorithm with an nlist of 4096 would take roughly 1.1 * (((4 * 128) * 2,000,000,000) + (4 * 4096 * 128)) bytes = 1126 GB.
This savings does come at a cost, however, because HNSW offers a better query latency versus approximation accuracy tradeoff.
Product quantization vector compression
Although you can use HNSW and IVF to speed up nearest neighbor search, they can consume a considerable amount of memory. When we get into the billion-vector scale, we start to require thousands of GBs of memory to support their index structures. As we scale up the number of vectors or the dimension of vectors, this requirement continues to grow. Is there a way to use noticeably less space for our k-NN index?
The answer is yes! In fact, there are a lot of different ways to reduce the amount of memory vectors require. You can change your embedding model to produce smaller vectors, or you can apply techniques like Principle Component Analysis (PCA) to reduce the vector’s dimensionality. Another approach is to use quantization. The general idea of vector quantization is to map a large vector space with continuous values into a smaller space with discrete values. When a vector is mapped into a smaller space, it requires fewer bits to represent. However, this comes at a cost—when mapping to a smaller input space, some information about the vector is lost.
Product quantization (PQ) is a very popular quantization technique in the field of nearest neighbor search. It can be used together with ANN algorithms for nearest neighbor search. Along with IVF, the k-NN plugin added support for Faiss’s PQ implementation in OpenSearch 1.2.
The main idea of PQ is to break up a vector into several sub-vectors and encode the sub-vectors independently with a fixed number of bits. The number of sub-vectors that the original vector is broken up into is controlled by a parameter, m, and the number of bits to encode each sub-vector with is controlled by a parameter, code_size. After encoding finishes, a vector is compressed into roughly m * code_size bits. So, assume we have a set of 100,000 1024-dimensional vectors. With m = 8 and code_size = 8, PQ breaks each vector into 8 128-dimensional sub-vectors and encode each sub-vector with 8 bits.
The values used for encoding are produced during a training step. During training, tables are created with 2code_size entries for each sub-vector partition. Next, k-Means clustering, with a k value of 2code_size, is run on the corresponding partition of sub-vectors from the training data. The centroids produced here are added as the entries to the partition’s table.
After all the tables are created, we encode a vector by replacing each sub-vector with the ID of the closest vector in the partition’s table. In the example where code_size = 8, we only need 8 bits to store an ID because there are 28 elements in the table. So, with dimension = 1024 and m = 8, the total size of one vector (assuming it uses a 32-bit floating point data type) is reduced from 4,096 bytes to roughly 8 bytes!

When we want to decode a vector, we can reconstruct an approximated version of it by using the stored IDs to retrieve the vectors from each partition’s table. The distance from the query vector to the reconstructed vector can then be computed and used in a nearest neighbor search. (It’s worth noting that, in practice, further optimization techniques like ADC are used to speed up this process for k-NN search).

Memory consumption
As we mentioned earlier, PQ will encode each vector into roughly m * code_size bits plus some overhead for each vector.
When combining it with IVF, we can estimate the index size as follows:
1.1 * ((((code_size/8) * m + overhead_per_vector) * num_vectors) + (4 * nlist * dimension) + (2 code_size * 4 * dimension) bytes
Using 1 billion vectors, dimension = 128, m = 8, code_size = 8, and nlist = 4096, we get an estimated total memory consumption of 70GB: 1.1 * ((((8 / 8) * 8 + 24) * 1,000,000,000) + (4 * 4096 * 128) + (2^8 * 4 * 128)) * 2 = 70 GB.
Running k-NN with OpenSearch
First make sure you have an OpenSearch cluster up and running. For instructions, refer to Cluster formation. For a more managed solution, you can use Amazon OpenSearch Service.
Before getting into the experiments, let’s go over how to run k-NN workloads in OpenSearch. First, we need to create an index. An index stores a set of documents in a way that they can be easily searched. For k-NN, the index’s mapping tells OpenSearch what algorithms to use and what parameters to use with them. We start by creating an index that uses HNSW as its search algorithm:
PUT my-hnsw-index
{
"settings": {
"index": {
"knn": true,
"number_of_shards": 10,
"number_of_replicas" 1,
}
},
"mappings": {
"properties": {
"my_vector": {
"type": "knn_vector",
"dimension": 4,
"method": {
"name": "hnsw",
"space_type": "l2",
"engine": "nmslib",
"parameters": {
"ef_construction": 128,
"m": 24
}
}
}
}
}
}
In the settings, we need to enable knn so that the index can be searched with the knn query type (more on this later). We also set the number of shards, and the number of replicas each shard will have. An index is made up of a collection of shards. Sharding is how OpenSearch distributes an index across multiple nodes in a cluster. For more information about shards, refer to Sizing Amazon OpenSearch Service domains.
In the mappings, we configure a field called my_vector of type knn_vector to store the vector data. We also pass nmslib as the engine to let OpenSearch know it should use nmslib’s implementation of HNSW. Additionally, we pass l2 as the space_type. The space_type determines the function used to compute the distance between two vectors. l2 refers to the Euclidean distance. OpenSearch also supports cosine similarity and the inner product distance functions.
After the index is created, we can ingest some fake data:
POST _bulk
{ "index": { "_index": "my-hnsw-index", "_id": "1" } }
{ "my_vector": [1.5, 2.5], "price": 12.2 }
{ "index": { "_index": "my-hnsw-index", "_id": "2" } }
{ "my_vector": [2.5, 3.5], "price": 7.1 }
{ "index": { "_index": "my-hnsw-index", "_id": "3" } }
{ "my_vector": [3.5, 4.5], "price": 12.9 }
{ "index": { "_index": "my-hnsw-index", "_id": "4" } }
{ "my_vector": [5.5, 6.5], "price": 1.2 }
{ "index": { "_index": "my-hnsw-index", "_id": "5" } }
{ "my_vector": [4.5, 5.5], "price": 3.7 }
{ "index": { "_index": "my-hnsw-index", "_id": "6" } }
{ "my_vector": [1.5, 5.5, 4.5, 6.4], "price": 10.3 }
{ "index": { "_index": "my-hnsw-index", "_id": "7" } }
{ "my_vector": [2.5, 3.5, 5.6, 6.7], "price": 5.5 }
{ "index": { "_index": "my-hnsw-index", "_id": "8" } }
{ "my_vector": [4.5, 5.5, 6.7, 3.7], "price": 4.4 }
{ "index": { "_index": "my-hnsw-index", "_id": "9" } }
{ "my_vector": [1.5, 5.5, 4.5, 6.4], "price": 8.9 }
After adding some documents to the index, we can search it:
GET my-hnsw-index/_search
{
"size": 2,
"query": {
"knn": {
"my_vector": {
"vector": [2, 3, 5, 6],
"k": 2
}
}
}
}
Creating an index that uses IVF or PQ is a little bit different because these algorithms require training. Before creating the index, we need to create a model using the training API:
POST /_plugins/_knn/models/my_ivfpq_model/_train
{
"training_index": "train-index",
"training_field": "train-field",
"dimension": 128,
"description": "My model description",
"method": {
"name":"ivf",
"engine":"faiss",
"parameters":{
"encoder":{
"name":"pq",
"parameters":{
"code_size": 8,
"m": 8
}
}
}
}
}
The training_index and training_field specify where the training data is stored. The only requirement for the training data index is that it has a knn_vector field that has the same dimension as you want your model to have. The method defines the algorithm that should be used for search.
After the training request is submitted, it will run in the background. To check if the training is complete, you can use the GET model API:
GET /_plugins/_knn/models/my_ivfpq_model/filter_path=model_id,state
{
"model_id" : "my_ivfpq_model",
"state" : "created"
}
After the model is created, you can create an index that uses this model:
PUT /my-hnsw-index
{
"settings" : {
"index.knn": true
"number_of_shards" : 10,
"number_of_replicas" : 1,
},
"mappings": {
"properties": {
"my_vector": {
"type": "knn_vector",
"model_id": "my_ivfpq_model"
}
}
}
}
After the index is created, we can add documents to it and search it just like we did for HNSW.
Experiments
Let’s run a few experiments to see how these algorithms perform in practice and what tradeoffs are made. We look at an HNSW versus an IVF index using PQ. For these experiments, we’re interested in search accuracy, query latency, and memory consumption. Because these trade-offs are mainly observed at scale, we use the BIGANN dataset containing 1 billion vectors of 128 dimensions. The dataset also contains 10,000 queries of test data mapping a query to the ground truth closest 100 vectors based on the Euclidean distance.
Specifically, we compute the following search metrics:
- Latency p99 (ms), Latency p90 (ms), Latency p50 (ms) – Query latency at various quantiles in milliseconds
- recall@10 – The fraction of the top 10 ground truth neighbors found in the 10 results returned by the plugin
- Native memory consumption (GB) – The amount of memory used by the plugin during querying
One thing to note is that the BIGANN dataset uses an unsigned integer as the data type. Because the knn_vector field doesn’t support unsigned integers, the data is automatically converted to floats.
To run the experiments, we complete the following steps:
- Ingest the dataset into the cluster using the OpenSearch Benchmarks framework (the code can be found on GitHub).
- When ingestion is complete, we use the warmup API to prepare the cluster for the search workload.
- We run the 10,000 test queries against the cluster 10 times and collect the aggregated results.
The queries return the document ID only, and not the vector, to improve performance (code for this can be found on GitHub).
Parameter selection
One tricky aspect of running experiments is selecting the parameters. There are too many different combinations of parameters to test them all. That being said, we decided to create three configurations for HNSW and IVFPQ:
- Optimize for search latency and memory
- Optimize for recall
- Fall somewhere in the middle
For each optimization strategy, we chose two configurations.
For HNSW, we can tune the m, ef_construction, and ef_search parameters to achieve our desired trade-off:
- m – Controls the maximum number of edges a node in a graph can have. Because each node has to store all of its edges, increasing this value will increase the memory footprint, but also increase the connectivity of the graph, which will improve recall.
- ef_construction – Controls the size of the candidate queue for edges when adding a node to the graph. Increasing this value will increase the number of candidates to consider, which will increase the index latency. However, because more candidates will be considered, the quality of the graph will be better, leading to better recall during search.
- ef_search – Similar to
ef_construction, it controls the size of the candidate queue for graph traversal during search. Increasing this value will increase the search latency, but will also improve the recall.
In general, we chose configurations that gradually increased the parameters, as detailed in the following table.
| Config ID |
Optimization Strategy |
m |
ef_construction |
ef_search |
| hnsw1 |
Optimize for memory and search latency |
8 |
32 |
32 |
| hnsw2 |
Optimize for memory and search latency |
16 |
32 |
32 |
| hnsw3 |
Balance between latency, memory, and recall |
16 |
128 |
128 |
| hnsw4 |
Balance between latency, memory, and recall |
32 |
256 |
256 |
| hnsw5 |
Optimize for recall |
32 |
512 |
512 |
| hnsw6 |
Optimize for recall |
64 |
512 |
512 |
For IVF, we can tune two parameters:
- nlist – Controls the granularity of the partitioning. The recommended value for this parameter is a function of the number of vectors in the index. One thing to keep in mind is that there are Faiss indexes that map to Lucene segments. There are several Lucene segments per shard and several shards per OpenSearch index. For our estimates, we assumed that there would be 100 segments per shard and 24 shards, so about 420,000 vectors per Faiss index. With this value, we estimated a good value to be 4096 and kept this constant for the experiments.
- nprobes – Controls the number of nlist buckets we search. Higher values generally lead to improved recalls at the expense of increased search latencies.
For PQ, we can tune two parameters:
- m – Controls the number of partitions to break the vector into. The larger this value is, the better the encoding will approximate the original, at the expense of raising memory consumption.
- code_size – Controls the number of bits to encode a sub-vector with. The larger this value is, the better the encoding approximates the original, at the expense of raising memory consumption. The max value is 8, so we kept it constant at 8 for all experiments.
The following table summarizes our strategies.
| Config ID |
Optimization Strategy |
nprobes |
m (num_sub_vectors) |
| ivfpq1 |
Optimize for memory and search latency |
8 |
8 |
| ivfpq2 |
Optimize for memory and search latency |
16 |
8 |
| ivfpq3 |
Balance between latency, memory, and recall |
32 |
16 |
| ivfpq4 |
Balance between latency, memory, and recall |
64 |
32 |
| ivfpq5 |
Optimize for recall |
128 |
16 |
| ivfpq6 |
Optimize for recall |
128 |
32 |
Additionally, we need to figure out how much training data to use for IVFPQ. In general, Faiss recommends between 30,000 and 256,000 training vectors for components involving k-Means training. For our configurations, the maximum k for k-Means is 4096 from the nlist parameter. With this formula, the recommended training set size is between 122,880 and 1,048,576 vectors, so we settled on 1 million vectors. The training data comes from the index vector dataset.
Lastly, for the index configurations, we need to select the shard count. It is recommended to keep the shard size between 10–50 GBs for OpenSearch. Experimentally, we determined that for HNSW, a good number would be 64 shards and for IVFPQ, 42. Both index configurations were configured with one replica.
Cluster configuration
To run these experiments, we used Amazon OpenSearch Service using version 1.3 of OpenSearch to create the clusters. We decided to use the r5 instance family, which provides a good trade-off between memory size and cost.
The number of nodes will depend on the amount of memory that can be used for the algorithm per node and the total amount of memory required by the algorithm. Having more nodes and more memory will generally improve performance, but for these experiments, we want to minimize cost. The amount of memory available per node is computed as memory_available = (node_memory - jvm_size) * circuit_breaker_limit, with the following parameters:
- node_memory – The total memory of the instance.
- jvm_size – The OpenSearch JVM heap size. Set to 32 GB.
- circuit_breaker_limit – The native memory usage threshold for the circuit breaker. Set to 0.5.
Because HNSW and IVFPQ have different memory requirements, we estimate how much memory is needed for each algorithm and determine the required number of nodes accordingly.
For HNSW, with m = 64, the total memory required using the formula from the previous sections is approximately 2,252 GB. Therefore, with r5.12xlarge (384 GB of memory), memory_available is 176 GB and the total number of nodes required is about 12, which we round up to 16 for stability purposes.
Because the IVFPQ algorithm requires less memory, we can use a smaller instance type, the r5.4xlarge instance, which has 128 GB of memory. Therefore, the memory_available for the algorithm is 48 GB. The estimated algorithm memory consumption where m = 64 is a total of 193 GB and the total number of nodes required is four, which we round up to six for stability purposes.
For both clusters, we use c5.2xlarge instance types as dedicated leader nodes. This will provide more stability for the cluster.
According to the AWS Pricing Calculator, for this particular use case, the cost per hour of the HNSW cluster is around $75 an hour, and the IVFPQ cluster costs around $11 an hour. This is important to remember when comparing the results.
Also, keep in mind that these benchmarks can be run using your custom infrastructure, using Amazon Elastic Compute Cloud (Amazon EC2), as long as the instance types and their memory size is equivalent.
Results
The following tables summarize the results from the experiments.
| Test ID |
p50 Query latency (ms) |
p90 Query latency (ms) |
p99 Query latency (ms) |
Recall@10 |
Native memory consumption (GB) |
| hnsw1 |
9.1 |
11 |
16.9 |
0.84 |
1182 |
| hnsw2 |
11 |
12.1 |
17.8 |
0.93 |
1305 |
| hnsw3 |
23.1 |
27.1 |
32.2 |
0.99 |
1306 |
| hnsw4 |
54.1 |
68.3 |
80.2 |
0.99 |
1555 |
| hnsw5 |
83.4 |
100.6 |
114.7 |
0.99 |
1555 |
| hnsw6 |
103.7 |
131.8 |
151.7 |
0.99 |
2055 |
| Test ID |
p50 Query latency (ms) |
p90 Query latency (ms) |
p99 Query latency (ms) |
Recall@10 |
Native memory consumption (GB) |
| ivfpq1 |
74.9 |
100.5 |
106.4 |
0.17 |
68 |
| ivfpq2 |
78.5 |
104.6 |
110.2 |
0.18 |
68 |
| ivfpq3 |
87.8 |
107 |
122 |
0.39 |
83 |
| ivfpq4 |
117.2 |
131.1 |
151.8 |
0.61 |
114 |
| ivfpq5 |
128.3 |
174.1 |
195.7 |
0.40 |
83 |
| ivfpq6 |
163 |
196.5 |
228.9 |
0.61 |
114 |
As you might expect, given how many more resources it uses, the HNSW cluster has lower query latencies and better recall. However, the IVFPQ indexes use significantly less memory.
For HNSW, increasing the parameters does in fact lead to better recall at the expense of latency. For IVFPQ, increasing m has the most significant impact on improving recall. Increasing nprobes improves the recall marginally, but at the expense of significant increases in latencies.
Conclusion
In this post, we covered different algorithms and techniques used to perform approximate k-NN search at scale (over 1 billion data points) within OpenSearch. As we saw in the previous benchmarks section, there isn’t one algorithm or approach that optimises for all the metrics at once. HNSW, IVF, and PQ each allow you to optimize for different metrics in your k-NN workload. When choosing the k-NN algorithm to use, first understand the requirements of your use case (How accurate does my approximate nearest neighbor search need to be? How fast should it be? What’s my budget?) and then tailor the algorithm configuration to meet them.
You can take a look at the benchmarking code base we used on GitHub. You can also get started with approximate k-NN search today following the instructions in Approximate k-NN search. If you’re looking for a managed solution for your OpenSearch cluster, check out Amazon OpenSearch Service.
About the Authors
 Jack Mazanec is a software engineer working on OpenSearch plugins. His primary interests include machine learning and search engines. Outside of work, he enjoys skiing and watching sports.
Jack Mazanec is a software engineer working on OpenSearch plugins. His primary interests include machine learning and search engines. Outside of work, he enjoys skiing and watching sports.
 Othmane Hamzaoui is a Data Scientist working at AWS. He is passionate about solving customer challenges using Machine Learning, with a focus on bridging the gap between research and business to achieve impactful outcomes. In his spare time, he enjoys running and discovering new coffee shops in the beautiful city of Paris.
Othmane Hamzaoui is a Data Scientist working at AWS. He is passionate about solving customer challenges using Machine Learning, with a focus on bridging the gap between research and business to achieve impactful outcomes. In his spare time, he enjoys running and discovering new coffee shops in the beautiful city of Paris.
 Mei Qu is a Business Intelligence Engineer on the AWS QuickSight ProServe Team. She helps customers and partners leverage their data to develop key business insights and monitor ongoing initiatives. She also develops QuickSight proof of concept projects, delivers technical workshops, and supports implementation projects.
Mei Qu is a Business Intelligence Engineer on the AWS QuickSight ProServe Team. She helps customers and partners leverage their data to develop key business insights and monitor ongoing initiatives. She also develops QuickSight proof of concept projects, delivers technical workshops, and supports implementation projects. Emily Zhu is a Senior Product Manager at Amazon QuickSight, AWS’s cloud-native, fully managed SaaS BI service. She leads the development of QuickSight analytics and query capabilities. Before joining AWS, she was working in the Amazon Prime Air drone delivery program and the Boeing company as senior strategist for several years. Emily is passionate about potentials of cloud-based BI solutions and looks forward to helping customers advance in their data-driven strategy-making.
Emily Zhu is a Senior Product Manager at Amazon QuickSight, AWS’s cloud-native, fully managed SaaS BI service. She leads the development of QuickSight analytics and query capabilities. Before joining AWS, she was working in the Amazon Prime Air drone delivery program and the Boeing company as senior strategist for several years. Emily is passionate about potentials of cloud-based BI solutions and looks forward to helping customers advance in their data-driven strategy-making.































 Jianwei Li is Senior Analytics Specialist TAM. He provides consultant service for AWS enterprise support customers to design and build modern data platform. He has more than 10 years experience in big data and analytics domain. In his spare time, he like running and hiking.
Jianwei Li is Senior Analytics Specialist TAM. He provides consultant service for AWS enterprise support customers to design and build modern data platform. He has more than 10 years experience in big data and analytics domain. In his spare time, he like running and hiking. Narayanan Venkateswaran is an Engineer in the AWS EMR group. He works on developing Hive in EMR. He has over 17 years of work experience in the industry across several companies including Sun Microsystems, Microsoft, Amazon and Oracle. Narayanan also holds a PhD in databases with focus on horizontal scalability in relational stores.
Narayanan Venkateswaran is an Engineer in the AWS EMR group. He works on developing Hive in EMR. He has over 17 years of work experience in the industry across several companies including Sun Microsystems, Microsoft, Amazon and Oracle. Narayanan also holds a PhD in databases with focus on horizontal scalability in relational stores. Partha Sarathi is an Analytics Specialist TAM – at AWS based in Sydney, Australia. He brings 15+ years of technology expertise and helps Enterprise customers optimize Analytics workloads. He has extensively worked on both on-premise and cloud Bigdata workloads along with various ETL platform in his previous roles. He also actively works on conducting proactive operational reviews around the Analytics services like Amazon EMR, Redshift, and OpenSearch.
Partha Sarathi is an Analytics Specialist TAM – at AWS based in Sydney, Australia. He brings 15+ years of technology expertise and helps Enterprise customers optimize Analytics workloads. He has extensively worked on both on-premise and cloud Bigdata workloads along with various ETL platform in his previous roles. He also actively works on conducting proactive operational reviews around the Analytics services like Amazon EMR, Redshift, and OpenSearch. Krish is an Enterprise Support Manager responsible for leading a team of specialists in EMEA focused on BigData & Analytics, Databases, Networking and Security. He is also an expert in helping enterprise customers modernize their data platforms and inspire them to implement operational best practices. In his spare time, he enjoys spending time with his family, travelling, and video games.
Krish is an Enterprise Support Manager responsible for leading a team of specialists in EMEA focused on BigData & Analytics, Databases, Networking and Security. He is also an expert in helping enterprise customers modernize their data platforms and inspire them to implement operational best practices. In his spare time, he enjoys spending time with his family, travelling, and video games.



















































































































 Jack Mazanec is a software engineer working on OpenSearch plugins. His primary interests include machine learning and search engines. Outside of work, he enjoys skiing and watching sports.
Jack Mazanec is a software engineer working on OpenSearch plugins. His primary interests include machine learning and search engines. Outside of work, he enjoys skiing and watching sports. Othmane Hamzaoui is a Data Scientist working at AWS. He is passionate about solving customer challenges using Machine Learning, with a focus on bridging the gap between research and business to achieve impactful outcomes. In his spare time, he enjoys running and discovering new coffee shops in the beautiful city of Paris.
Othmane Hamzaoui is a Data Scientist working at AWS. He is passionate about solving customer challenges using Machine Learning, with a focus on bridging the gap between research and business to achieve impactful outcomes. In his spare time, he enjoys running and discovering new coffee shops in the beautiful city of Paris.