Post Syndicated from Channy Yun original https://aws.amazon.com/blogs/aws/aws-weekly-roundup-aws-chips-taste-test-generative-ai-updates-community-days-and-more-april-1-2024/
Today is April Fool’s Day. About 10 years ago, some tech companies would joke about an idea that was thought to be fun and unfeasible on April 1st, to the delight of readers. Jeff Barr has also posted seemingly far-fetched ideas on this blog in the past, and some of these have surprisingly come true! Here are examples:
| Year | Joke | Reality |
| 2010 | Introducing QC2 – the Quantum Compute Cloud, a production-ready quantum computer to solve certain types of math and logic problems with breathtaking speed. | In 2019, we launched Amazon Braket, a fully managed service that allows scientists, researchers, and developers to begin experimenting with computers from multiple quantum hardware providers in a single place. |
| 2011 | Announcing AWS $NAME, a scalable event service to find and automatically integrate with your systems on the cloud, on premises, and even your house and room. | In 2019, we introduced Amazon EventBridge to make it easy for you to integrate your own AWS applications with third-party applications. If you use AWS IoT Events, you can monitor and respond to events at scale from your IoT devices at home. |
| 2012 | New Amazon EC2 Fresh Servers to deliver a fresh (physical) EC2 server in 15 minutes using atmospheric delivery and communucation from a fleet of satellites. | In 2021, we launched AWS Outposts Server, 1U/2U physical servers with built-in AWS services. In 2023, Project Kuiper completed successful tests of an optical mesh network in low Earth orbit. Now, we only need to develop satellite warehouse and atmospheric re-entry technology to follow Amazon PrimeAir’s drone delivery. |
| 2013 | PC2 – The New Punched Card Cloud, a new mf (mainframe) instance family, Mainframe Machine Images (MMI), tape storage, and punched card interfaces for mainframe computers used from the 1970s to ’80s. | In 2022, we launched AWS Mainframe Modernization to help you modernize your mainframe applications and deploy them to AWS fully managed runtime environments. |
 Jeff returns! This year, we have AWS “Chips” Taste Test for him to indulge in, drawing unique parallels between chip flavors and silicon innovations. He compared the taste of “Golden Nacho Cheese,” “Al Chili Lime,” and “BBQ Training Wheels” with AWS Graviton, AWS Inferentia, and AWS Trainium chips.
Jeff returns! This year, we have AWS “Chips” Taste Test for him to indulge in, drawing unique parallels between chip flavors and silicon innovations. He compared the taste of “Golden Nacho Cheese,” “Al Chili Lime,” and “BBQ Training Wheels” with AWS Graviton, AWS Inferentia, and AWS Trainium chips.
What’s your favorite? Watch a fun video in the LinkedIn and X post of AWS social media channels.
Last week’s launches
If we stay curious, keep learning, and insist on high standards, we will continue to see more ideas turn into reality. The same goes for the generative artificial intelligence (generative AI) world. Here are some launches that utilize generative AI technology this week.
Knowledge Bases for Amazon Bedrock – Anthropic’s Claude 3 Sonnet foundation model (FM) is now generally available on Knowledge Bases for Amazon Bedrock to connect internal data sources for Retrieval Augmented Generation (RAG).
Knowledge Bases for Amazon Bedrock support metadata filtering, which improves retrieval accuracy by ensuring the documents are relevant to the query. You can narrow search results by specifying which documents to include or exclude from a query, resulting in more relevant responses generated by FMs such as Claude 3 Sonnet.

Finally, you can customize prompts and number of retrieval results in Knowledge Bases for Amazon Bedrock. With custom prompts, you can tailor the prompt instructions by adding context, user input, or output indicator(s), for the model to generate responses that more closely match your use case needs. You can now control the amount of information needed to generate a final response by adjusting the number of retrieved passages. To learn more these new features, visit Knowledge bases for Amazon Bedrock in the AWS documentation.
Amazon Connect Contact Lens – At AWS re:Invent 2023, we previewed a generative AI capability to summarize long customer conversations into succinct, coherent, and context-rich contact summaries to help improve contact quality and agent performance. These generative AI–powered post-contact summaries are now available in Amazon Connect Contact Lens.

Amazon DataZone – At AWS re:Invent 2023, we also previewed a generative AI–based capability to generate comprehensive business data descriptions and context and include recommendations on analytical use cases. These generative AI–powered recommendations for descriptions are now available in Amazon DataZone.

There are also other important launches you shouldn’t miss:
A new Local Zone in Miami, Florida – AWS Local Zones are an AWS infrastructure deployment that places compute, storage, database, and other select services closer to large populations, industry, and IT centers where no AWS Region exists. You can now use a new Local Zone in Miami, Florida, to run applications that require single-digit millisecond latency, such as real-time gaming, hybrid migrations, and live video streaming. Enable the new Local Zone in Miami (use1-mia2-az1) from the Zones tab in the Amazon EC2 console settings to get started.

New Amazon EC2 C7gn metal instance – You can use AWS Graviton based new C7gn bare metal instances to run applications that benefit from deep performance analysis tools, specialized workloads that require direct access to bare metal infrastructure, legacy workloads not supported in virtual environments, and licensing-restricted business-critical applications. The EC2 C7gn metal size comes with 64 vCPUs and 128 GiB of memory.
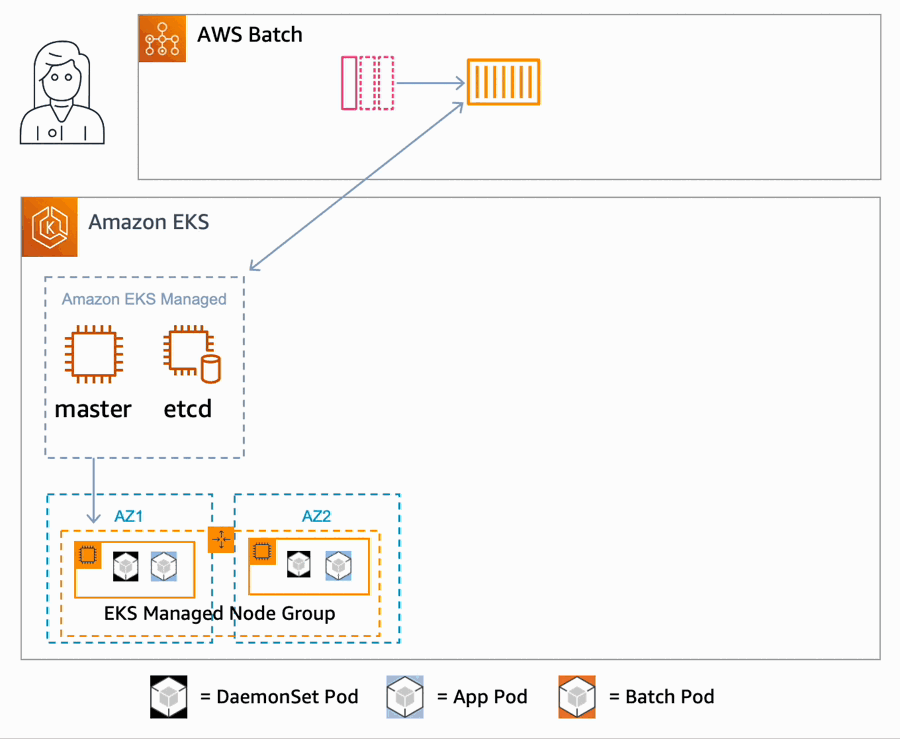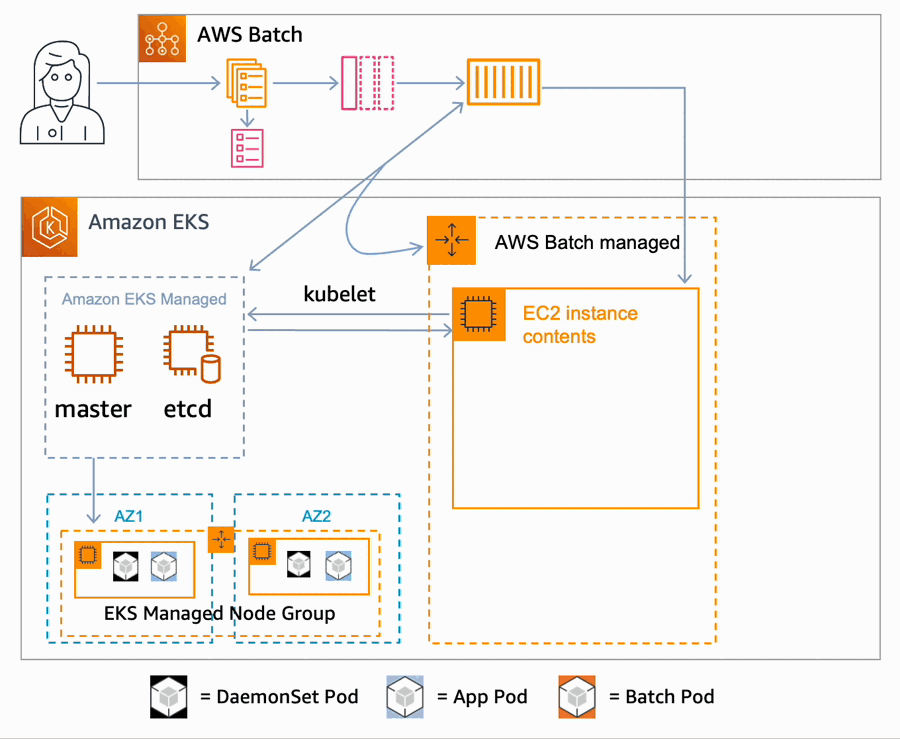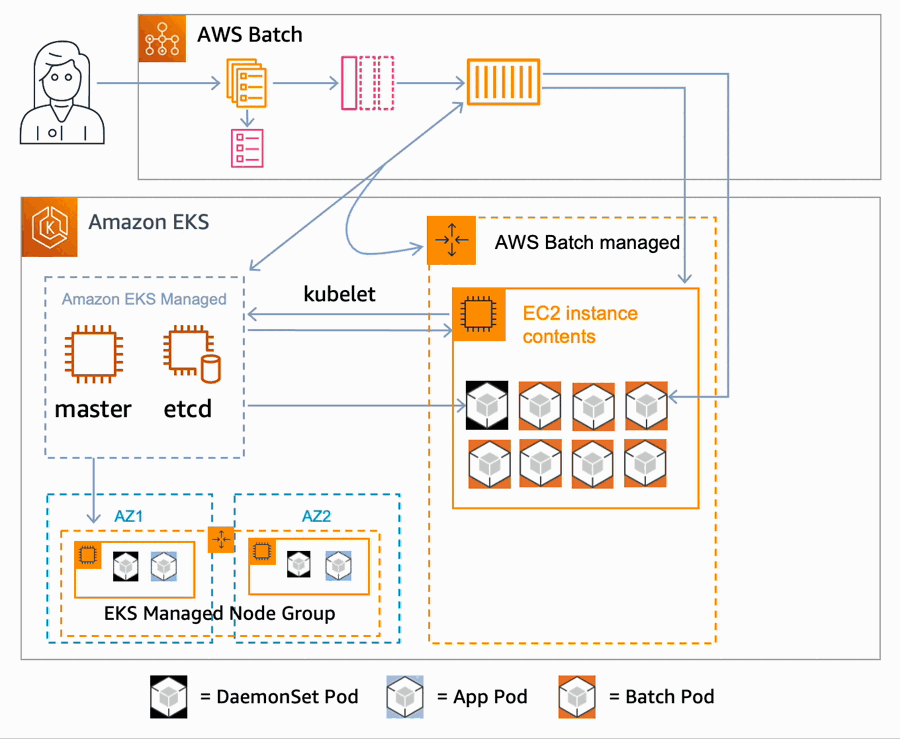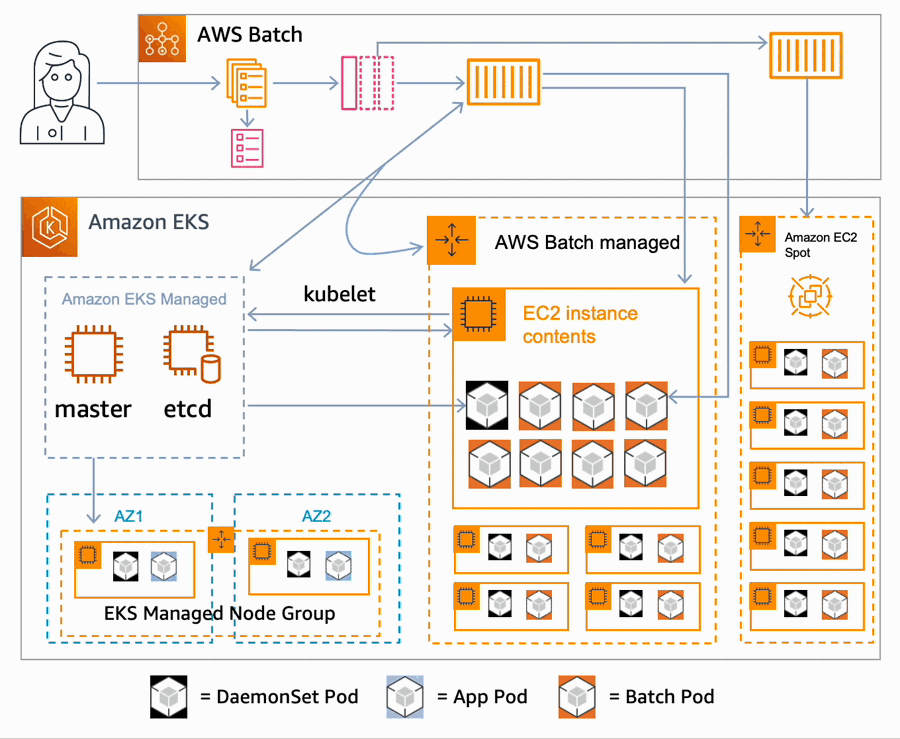
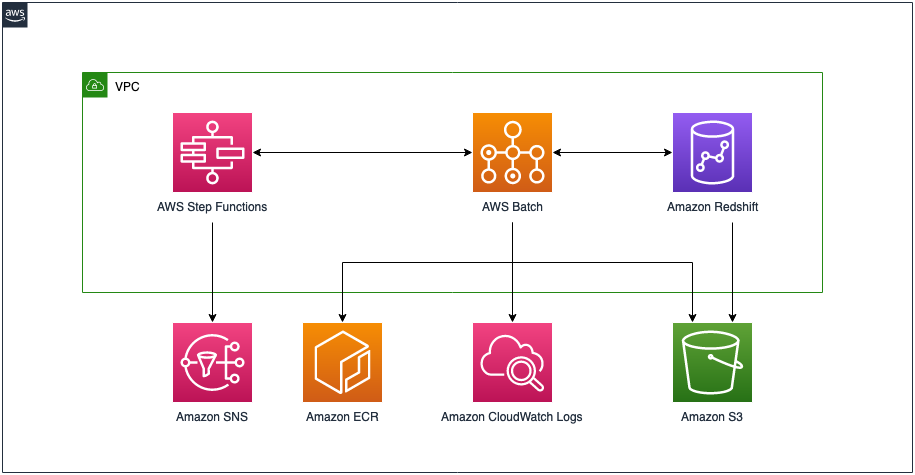
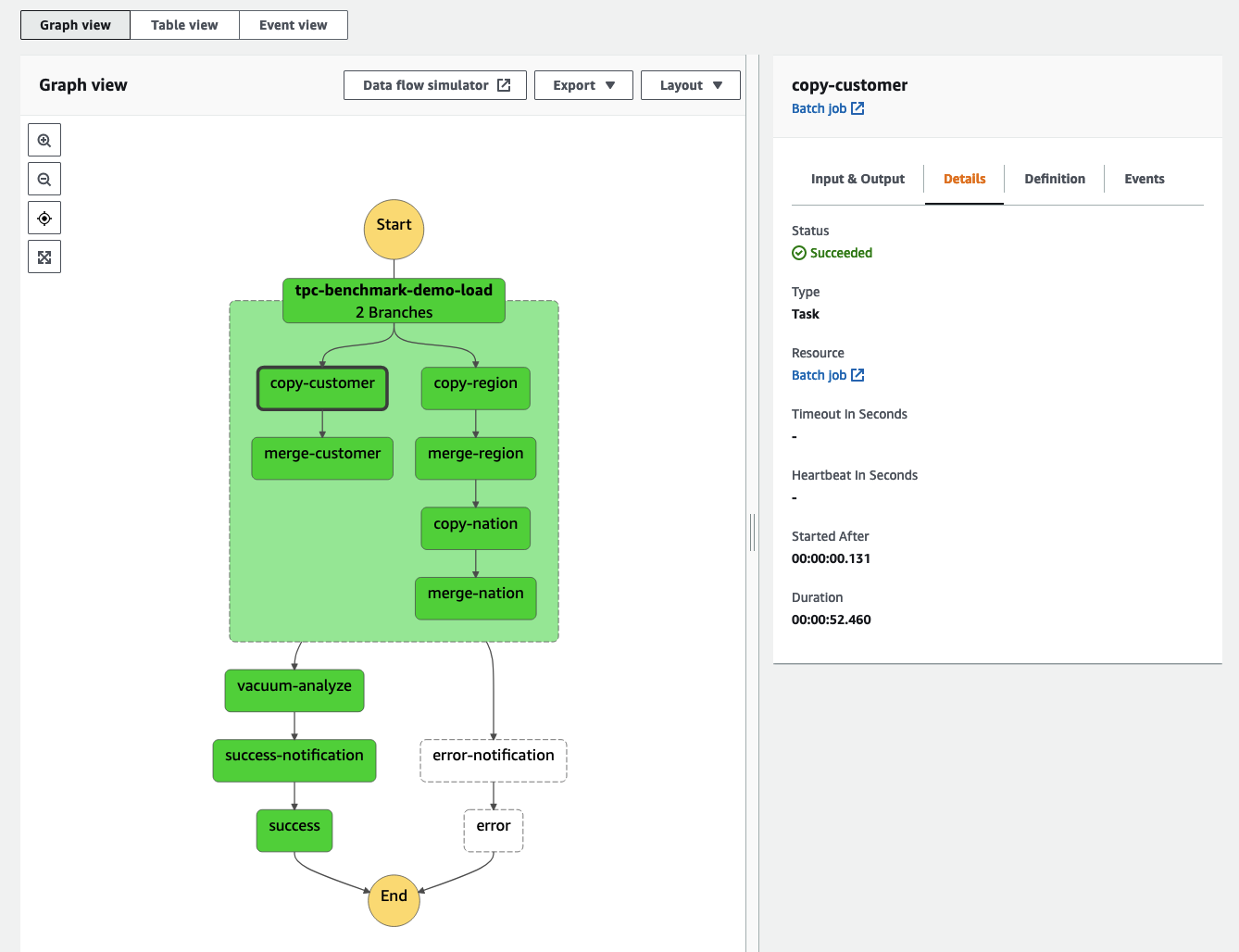
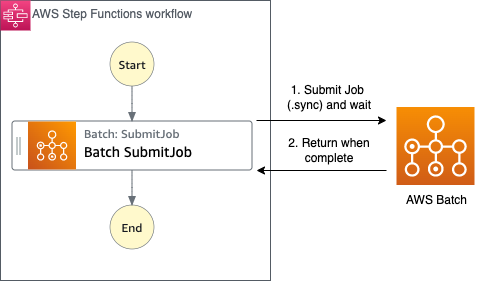
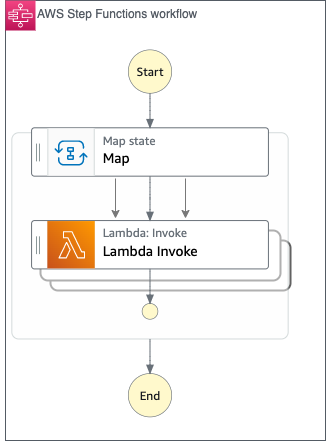
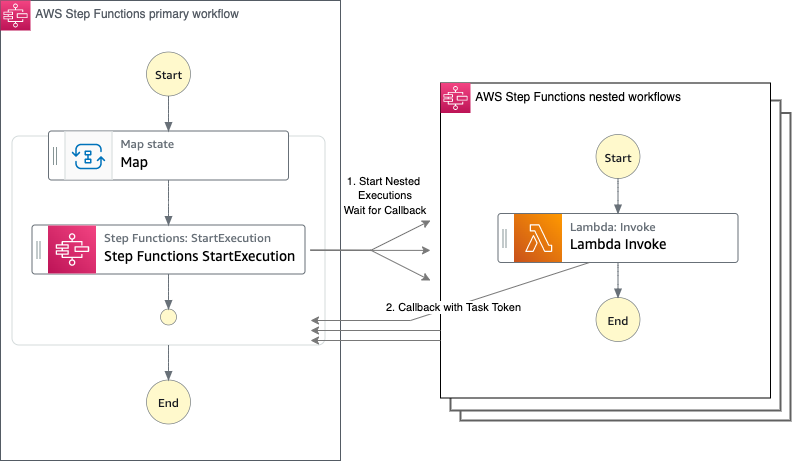
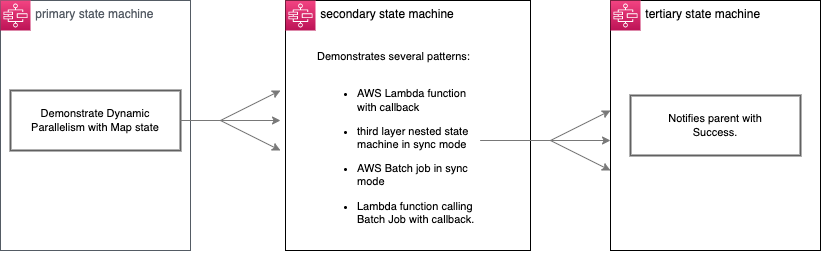
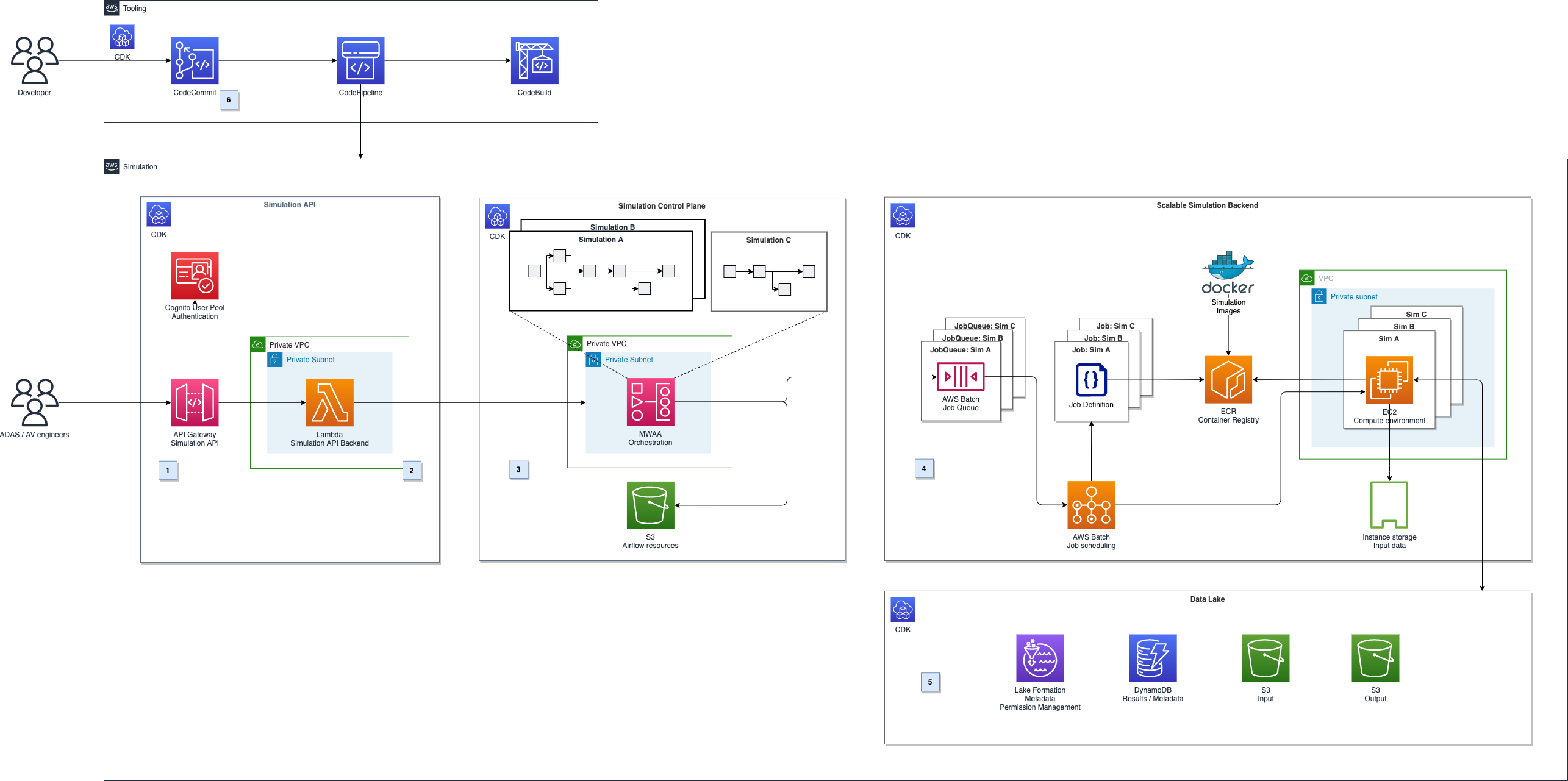
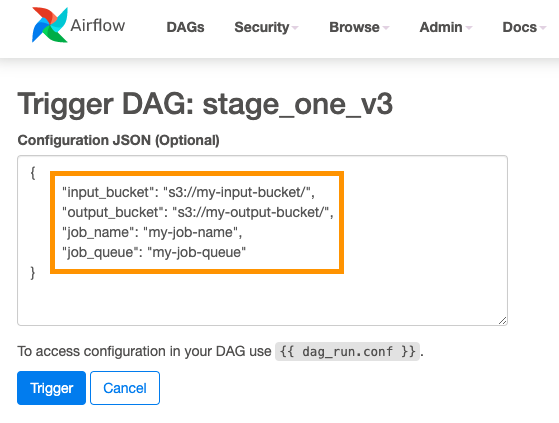
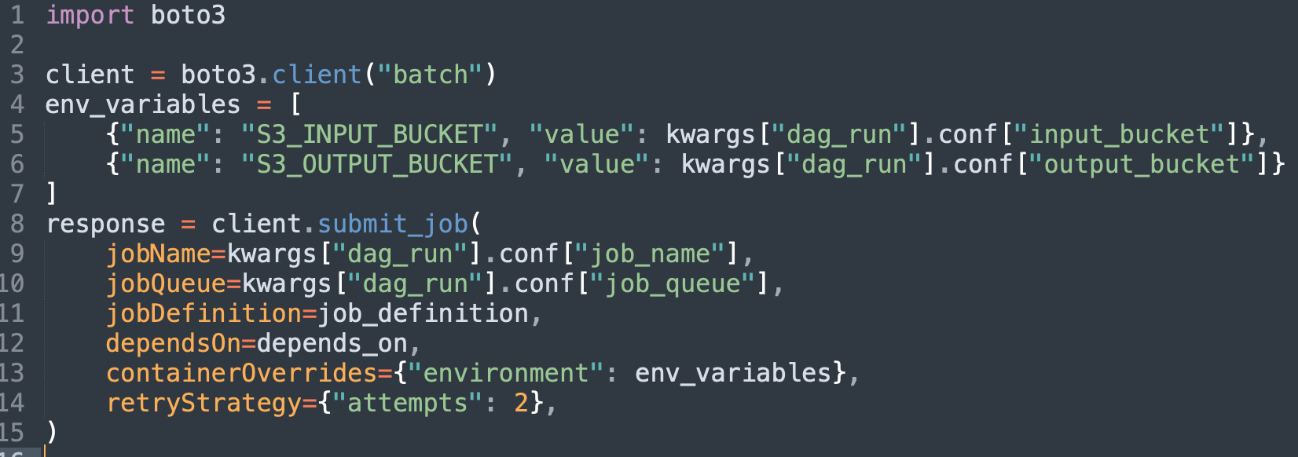
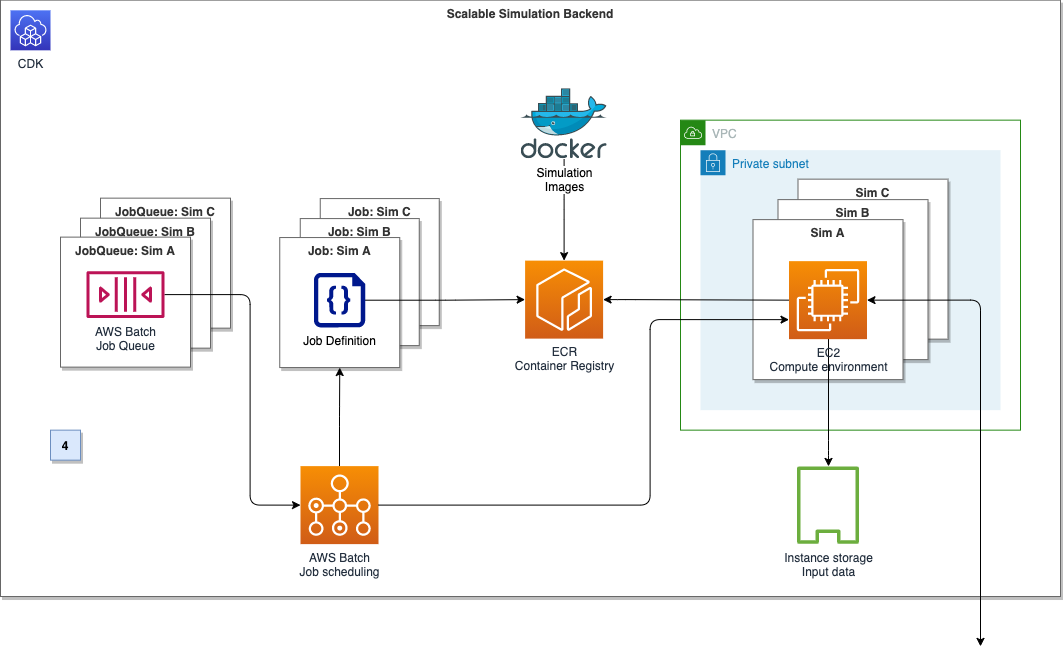
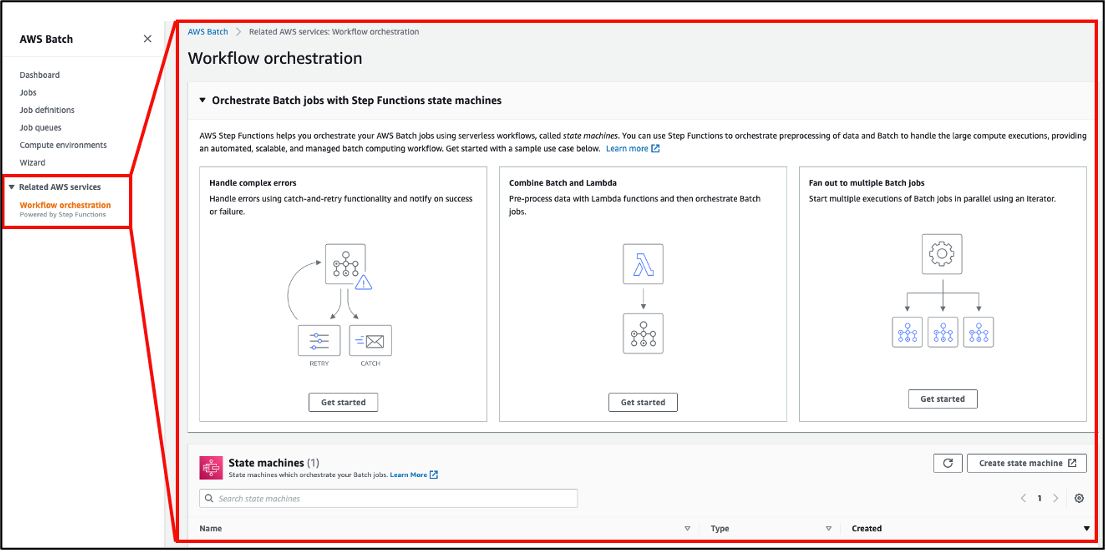
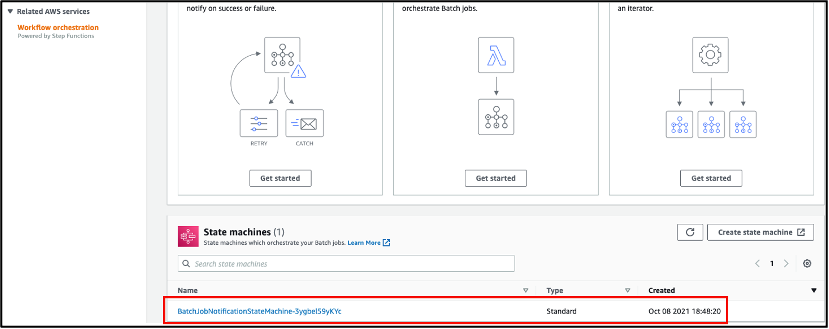
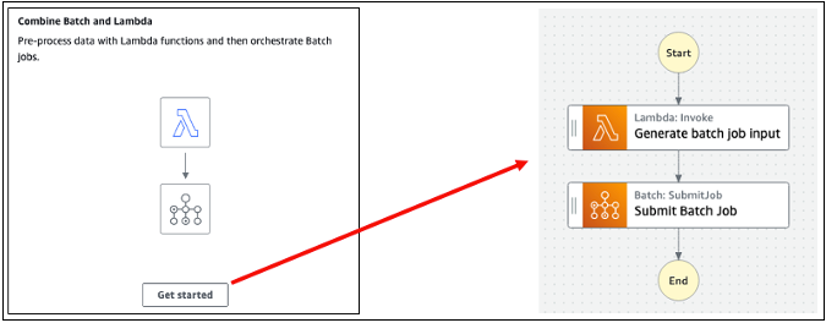
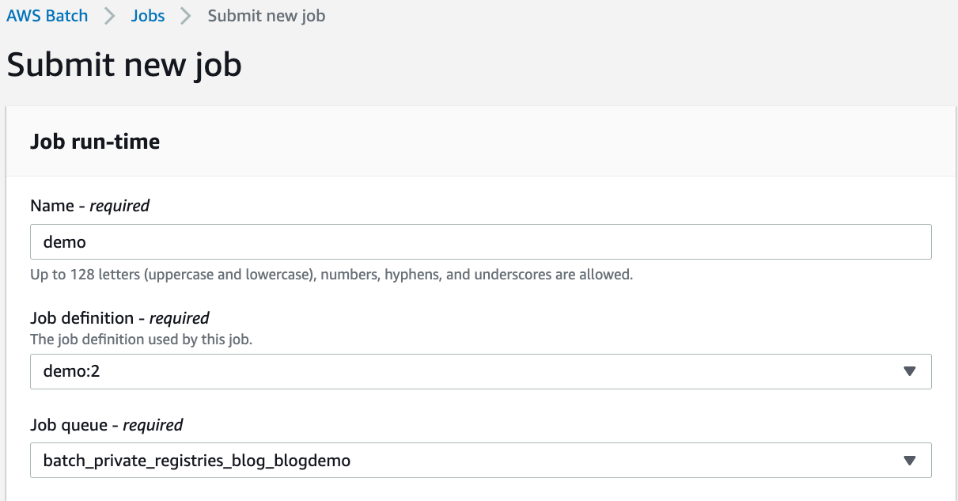
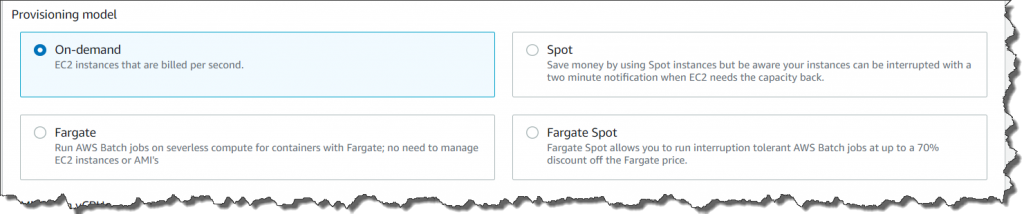
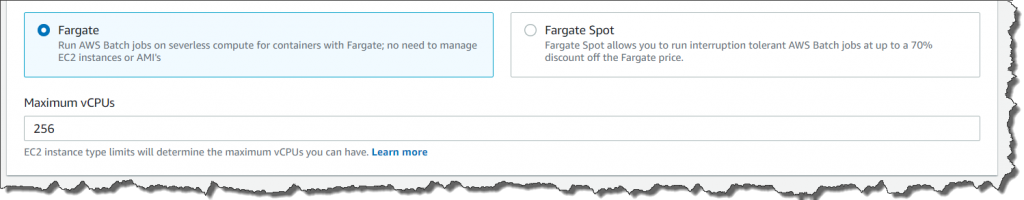
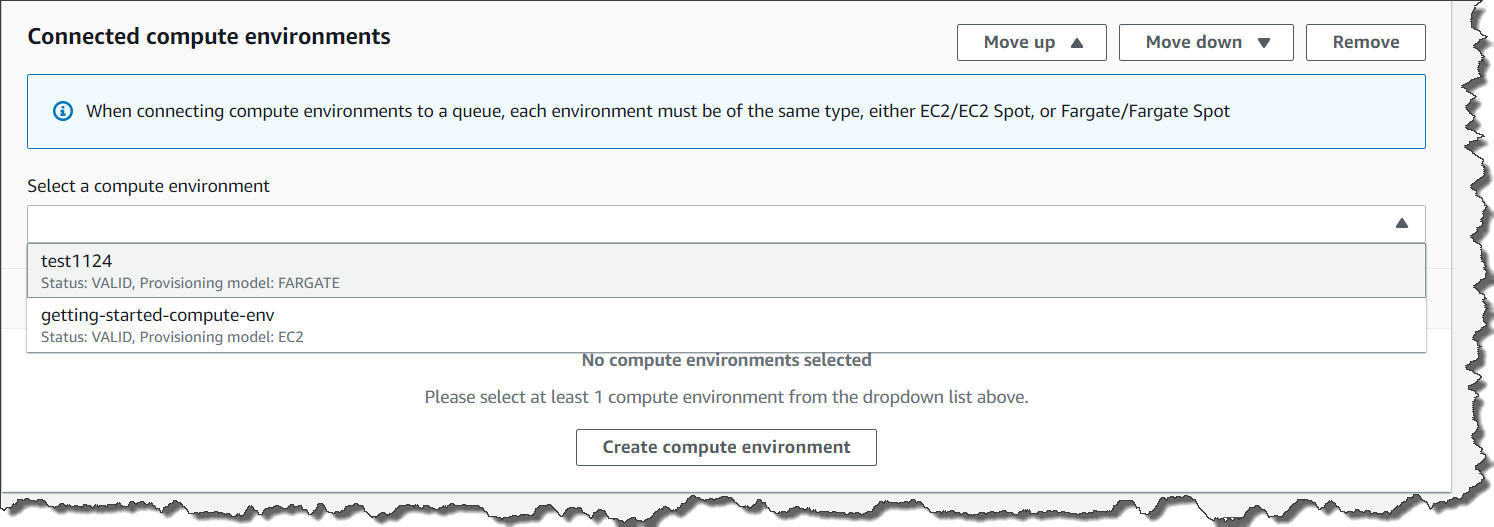
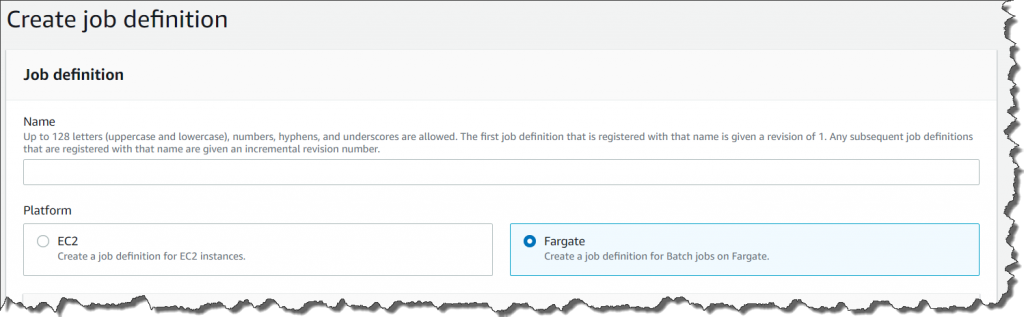
AWS Batch multi-container jobs – You can use multi-container jobs in AWS Batch, making it easier and faster to run large-scale simulations in areas like autonomous vehicles and robotics. With the ability to run multiple containers per job, you get the advanced scaling, scheduling, and cost optimization offered by AWS Batch, and you can use modular containers representing different components like 3D environments, robot sensors, or monitoring sidecars.
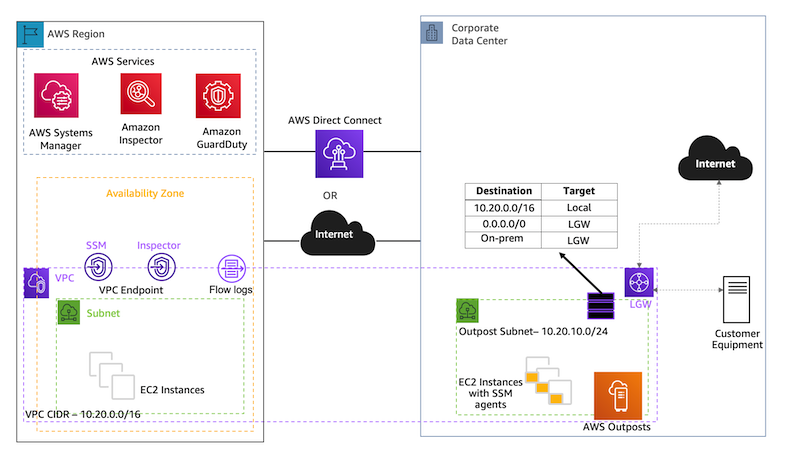
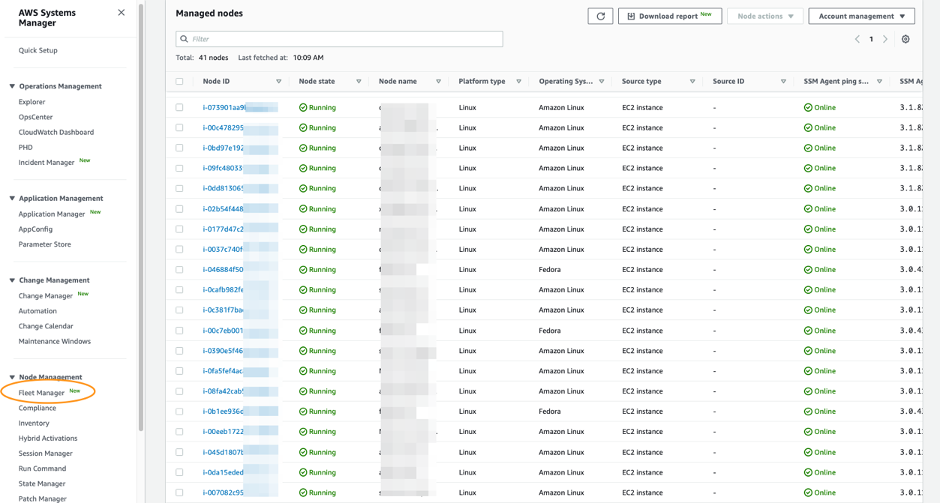
Amazon Guardduty EC2 Runtime Monitoring – We are announcing the general availability of Amazon GuardDuty EC2 Runtime Monitoring to expand threat detection coverage for EC2 instances at runtime and complement the anomaly detection that GuardDuty already provides by continuously monitoring VPC Flow Logs, DNS query logs, and AWS CloudTrail management events. You now have visibility into on-host, OS-level activities and container-level context into detected threats.
GitLab support for AWS CodeBuild – You can now use GitLab and GitLab self-managed as the source provider for your CodeBuild projects. You can initiate builds from changes in source code hosted in your GitLab repositories. To get started with CodeBuild’s new source providers, visit the AWS CodeBuild User Guide.
Retroactive support for AWS cost allocation tags – You can enable AWS cost allocation tags retroactively for up to 12 months. Previously, when you activated resource tags for cost allocation purposes, the tags only took effect prospectively. Submit a backfill request, specifying the duration of time you want the cost allocation tags to be backfilled. Once the backfill is complete, the cost and usage data from prior months will be tagged with the current cost allocation tags.
For a full list of AWS announcements, be sure to keep an eye on the What’s New at AWS page.
Other AWS News
Some other updates and news about generative AI that you might have missed:
Amazon and Anthropic’s AI investiment – Read the latest milestone in our strategic collaboration and investment with Anthropic. Now, Anthropic is using AWS as its primary cloud provider and will use AWS Trainium and Inferentia chips for mission-critical workloads, including safety research and future FM development. Earlier this month, we announced access to Anthropic’s most powerful FM, Claude 3, on Amazon Bedrock. We announced availability of Sonnet on March 4 and Haiku on March 13. To learn more, watch the video introducing Claude on Amazon Bedrock.
Virtual building assistant built on Amazon Bedrock – BrainBox AI announced the launch of ARIA (Artificial Responsive Intelligent Assistant) powered by Amazon Bedrock. ARIA is designed to enhance building efficiency by assimilating seamlessly into the day-to-day processes related to building management. To learn more, read the full customer story and watch the video on how to reduce a building’s CO2 footprint with ARIA.
Solar models available on Amazon SageMaker JumpStart – Upstage Solar is a large language model (LLM) 100 percent pre-trained with Amazon SageMaker that outperforms and uses its compact size and powerful track record to specialize in purpose training, making it versatile across languages, domains, and tasks. Now, Solar Mini is available on Amazon SageMaker JumpStart. To learn more, watch how to deploy Solar models in SageMaker JumpStart.
AWS open source news and updates – My colleague Ricardo writes this weekly open source newsletter in which he highlights new open source projects, tools, and demos from the AWS Community. Last week’s highlight was news that Linux Foundation launched Valkey community, an open source alternative to the Redis in-memory, NoSQL data store.
Upcoming AWS Events
Check your calendars and sign up for upcoming AWS events:
![]() AWS Summits – Join free online and in-person events that bring the cloud computing community together to connect, collaborate, and learn about AWS. Register in your nearest city: Paris (April 3), Amsterdam (April 9), Sydney (April 10–11), London (April 24), Berlin (May 15–16), and Seoul (May 16–17), Hong Kong (May 22), Milan (May 23), Dubai (May 29), Stockholm (June 4), and Madrid (June 5).
AWS Summits – Join free online and in-person events that bring the cloud computing community together to connect, collaborate, and learn about AWS. Register in your nearest city: Paris (April 3), Amsterdam (April 9), Sydney (April 10–11), London (April 24), Berlin (May 15–16), and Seoul (May 16–17), Hong Kong (May 22), Milan (May 23), Dubai (May 29), Stockholm (June 4), and Madrid (June 5).
![]() AWS re:Inforce – Explore cloud security in the age of generative AI at AWS re:Inforce, June 10–12 in Pennsylvania for two-and-a-half days of immersive cloud security learning designed to help drive your business initiatives. Read the story from AWS Chief Information Security Officer (CISO) Chris Betz about a bit of what you can expect at re:Inforce.
AWS re:Inforce – Explore cloud security in the age of generative AI at AWS re:Inforce, June 10–12 in Pennsylvania for two-and-a-half days of immersive cloud security learning designed to help drive your business initiatives. Read the story from AWS Chief Information Security Officer (CISO) Chris Betz about a bit of what you can expect at re:Inforce.
![]() AWS Community Days – Join community-led conferences that feature technical discussions, workshops, and hands-on labs led by expert AWS users and industry leaders from around the world: Mumbai (April 6), Poland (April 11), Bay Area (April 12), Kenya (April 20), and Turkey (May 18).
AWS Community Days – Join community-led conferences that feature technical discussions, workshops, and hands-on labs led by expert AWS users and industry leaders from around the world: Mumbai (April 6), Poland (April 11), Bay Area (April 12), Kenya (April 20), and Turkey (May 18).
You can browse all upcoming AWS led in-person and virtual events and developer-focused events such as AWS DevDay.
That’s all for this week. Check back next Monday for another Week in Review!
— Channy